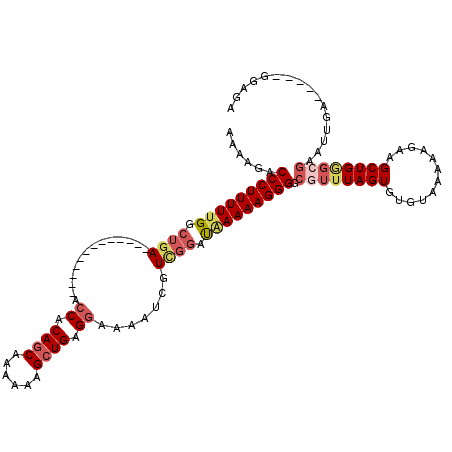
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,385,367 – 16,385,469 |
| Length | 102 |
| Max. P | 0.946022 |

| Location | 16,385,367 – 16,385,469 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -18.28 |
| Consensus MFE | -13.72 |
| Energy contribution | -14.13 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16385367 102 + 22407834 UCUCC-----ACAAUUCGCCCAGCUUCUUUUUACACACUAAACGCCCCUUUUUAUCCGACGAUUUUCCUCAGCUUUUUUGCUGUGGU-------------UCAGCCAAAAAGGGUCUUUU .....-----.................................(.((((((((....(..((....((.((((......)))).)).-------------))..).)))))))).).... ( -16.50) >DroSec_CAF1 8900 102 + 1 UCUCC-----UCAACUCGCCCAGCUUCUUUUUACACACUAAACGCCCCUUUUUAUCCGACGAUUUUCCUCAGCUUUUUUGCUGUGGU-------------UCAGCCAAAAAGGGUCUUUU .....-----.................................(.((((((((....(..((....((.((((......)))).)).-------------))..).)))))))).).... ( -16.50) >DroSim_CAF1 7014 102 + 1 UCUGC-----UCAAUUCUCCCAGCUUCUUUUUACACACUAAACGCCCCUUUUUAUCCGACGAUUUUCCUCAGCUUUUUUGCUGUGGU-------------UCAGCCAAAAAGGGUCUUUU ...((-----(..........)))...................(.((((((((....(..((....((.((((......)))).)).-------------))..).)))))))).).... ( -16.80) >DroEre_CAF1 7056 104 + 1 UCUCUUCUCC--AUUUCGCCCAGCUUCUUUUUACACACUAAACGCCCCUUUUUAUCCAAGGAUUUUCCUCAGCUU-UUUGCUGUGGC-------------UCAGCCAAAAAGGGUCUUUU ..........--...............................(.((((((((.....(((.....)))((((..-...)))).(((-------------...))))))))))).).... ( -18.10) >DroYak_CAF1 10242 107 + 1 UCUCCUCUCCUCAACUCGCUCAGCUUCUUUUUACACACUAAACGCCCCUUUUCAUCCAACGAUUUUCCUCAGCUUUUUUGCUGUGCU-------------UCAGCCAAAAAGGGUCUUUU ...........................................(.(((((((.................((((......)))).((.-------------...))..))))))).).... ( -13.00) >DroAna_CAF1 6443 101 + 1 -------------------GCAGCUUCUUUUUCGACACUAAACGCCCCUUUUUGUGCGAUGAUUUUCCUCAGCUUUUUCGAUGUGGUGGGUGGGUGGUGCUUCGGCGAAAAGGGUCCUUG -------------------..((..((((((((...((((..(((((((..(((.((((.(.....).)).)).....)))...)).)))))..))))((....))))))))))..)).. ( -28.80) >consensus UCUCC_____UCAAUUCGCCCAGCUUCUUUUUACACACUAAACGCCCCUUUUUAUCCGACGAUUUUCCUCAGCUUUUUUGCUGUGGU_____________UCAGCCAAAAAGGGUCUUUU ...........................................(.(((((((((((....)))...((.((((......)))).))....................)))))))).).... (-13.72 = -14.13 + 0.42)

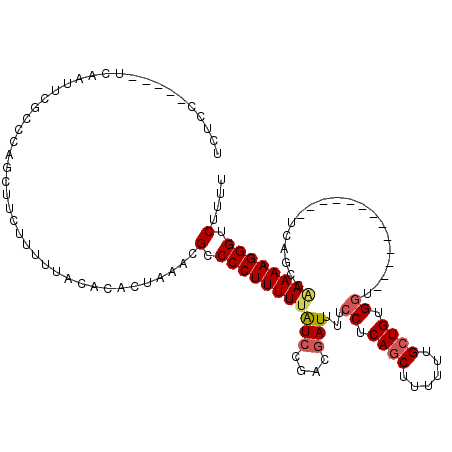
| Location | 16,385,367 – 16,385,469 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -25.45 |
| Consensus MFE | -20.21 |
| Energy contribution | -21.52 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16385367 102 - 22407834 AAAAGACCCUUUUUGGCUGA-------------ACCACAGCAAAAAAGCUGAGGAAAAUCGUCGGAUAAAAAGGGGCGUUUAGUGUGUAAAAAGAAGCUGGGCGAAUUGU-----GGAGA ......(((((((((.((((-------------.((.((((......)))).)).......)))).))))))))).((((((((............))))))))......-----..... ( -28.50) >DroSec_CAF1 8900 102 - 1 AAAAGACCCUUUUUGGCUGA-------------ACCACAGCAAAAAAGCUGAGGAAAAUCGUCGGAUAAAAAGGGGCGUUUAGUGUGUAAAAAGAAGCUGGGCGAGUUGA-----GGAGA ......(((((((((.((((-------------.((.((((......)))).)).......)))).))))))))).((((((((............))))))))......-----..... ( -28.50) >DroSim_CAF1 7014 102 - 1 AAAAGACCCUUUUUGGCUGA-------------ACCACAGCAAAAAAGCUGAGGAAAAUCGUCGGAUAAAAAGGGGCGUUUAGUGUGUAAAAAGAAGCUGGGAGAAUUGA-----GCAGA ......(((((((((.((((-------------.((.((((......)))).)).......)))).)))))))))..(((((((.....................)))))-----))... ( -25.10) >DroEre_CAF1 7056 104 - 1 AAAAGACCCUUUUUGGCUGA-------------GCCACAGCAAA-AAGCUGAGGAAAAUCCUUGGAUAAAAAGGGGCGUUUAGUGUGUAAAAAGAAGCUGGGCGAAAU--GGAGAAGAGA ......(((((((((((((.-------------....))))...-...(..(((.....)))..).))))))))).((((((((............))))))))....--.......... ( -29.50) >DroYak_CAF1 10242 107 - 1 AAAAGACCCUUUUUGGCUGA-------------AGCACAGCAAAAAAGCUGAGGAAAAUCGUUGGAUGAAAAGGGGCGUUUAGUGUGUAAAAAGAAGCUGAGCGAGUUGAGGAGAGGAGA ......(((((((..(((..-------------.((.((((......))))......(((....)))........))(((((((............))))))).)))..))))).))... ( -26.20) >DroAna_CAF1 6443 101 - 1 CAAGGACCCUUUUCGCCGAAGCACCACCCACCCACCACAUCGAAAAAGCUGAGGAAAAUCAUCGCACAAAAAGGGGCGUUUAGUGUCGAAAAAGAAGCUGC------------------- ......(((((((.((....))............((.((.(......).)).))..............))))))).....((((.((......)).)))).------------------- ( -14.90) >consensus AAAAGACCCUUUUUGGCUGA_____________ACCACAGCAAAAAAGCUGAGGAAAAUCGUCGGAUAAAAAGGGGCGUUUAGUGUGUAAAAAGAAGCUGGGCGAAUUGA_____GGAGA ......(((((((((.((((..............((.((((......)))).)).......)))).))))))))).((((((((............))))))))................ (-20.21 = -21.52 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:06 2006