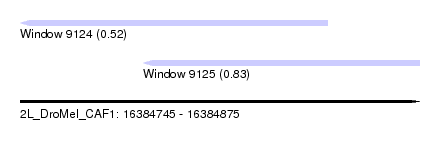
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,384,745 – 16,384,875 |
| Length | 130 |
| Max. P | 0.830173 |

| Location | 16,384,745 – 16,384,845 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 92.20 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -18.40 |
| Energy contribution | -20.04 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
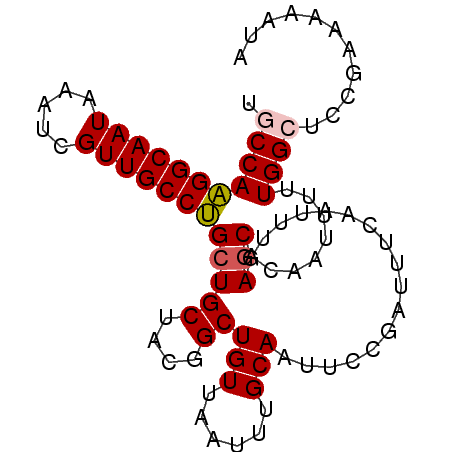
>2L_DroMel_CAF1 16384745 100 - 22407834 ACGGCUGUUAAUUUGCAAUUCCGAUUUCAAUUUGAGCACAAUUUUUGGAUCCGAAAAAUAUGGCCAACGGGCAAUUGCUCUGCAAUGCCUGUUGUACACU .(((.(((......)))...)))..................((((((....))))))...((..(((((((((.((((...)))))))))))))..)).. ( -25.90) >DroSec_CAF1 8279 100 - 1 ACGGCUGUUAAUUUGCAAUUCCGAUUUCAAUUUGAGCACAAUUUUUGGCUCUGAAAAAUAUGGCCAACGGGCAAUUGCUCUGCAAUGCCUGUUGUACACU .(((.(((......)))...))).(((((....((((.((.....)))))))))))....((..(((((((((.((((...)))))))))))))..)).. ( -29.60) >DroSim_CAF1 6393 100 - 1 ACGGCUGUUAAUUUGCAAUUCCGAUUUCAAUUUGAGCACAAUUUUUGGCUCCGAAAAAUAUGGCCAACGGGCAAUUGCUCUGCAAUGCCUGUUGUACACU .(((.(((......)))...)))..........((((.((.....)))))).........((..(((((((((.((((...)))))))))))))..)).. ( -27.60) >DroEre_CAF1 6465 89 - 1 ACGGCUGUUAAUUUGCAAUUCCGAUUUCAAUUUGAGCACAGUUUUUGGCUCCGAAAAAUAUGGCCAACGGGCAAUUGC-----------UGUUAUGCACU ...((...((((..(((((((((....)...(((.((.((.((((((....))))))...)))))))..)).))))))-----------.)))).))... ( -19.10) >DroYak_CAF1 9627 100 - 1 ACGGCUGUUAAUUUGCAAUUCCUAUUUCAAUUUGAGCACAAUUUUUGGCUCCGAAAAAUUUGGCCAACGGGCAAUUGCUCUGCAAUGCCUGCUAUACACU ..((((.((((.(((.(((....))).))).))))......((((((....))))))....))))..((((((.((((...))))))))))......... ( -22.70) >consensus ACGGCUGUUAAUUUGCAAUUCCGAUUUCAAUUUGAGCACAAUUUUUGGCUCCGAAAAAUAUGGCCAACGGGCAAUUGCUCUGCAAUGCCUGUUGUACACU ..((((.((((.(((.(((....))).))).))))......((((((....))))))....))))((((((((.((((...))))))))))))....... (-18.40 = -20.04 + 1.64)


| Location | 16,384,785 – 16,384,875 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -20.66 |
| Consensus MFE | -18.36 |
| Energy contribution | -18.64 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16384785 90 - 22407834 UGCCAAGGCAAUAAAUCGUUGCCUGCUGCUACGGCUGUUAAUUUGCAAUUCCGAUUUCAAUUUGAGCACAAUUUUUGGAUCCGAAAAAUA (((..(((((((.....)))))))((.((....)).))......))).(((.((((.(((.(((....)))...))))))).)))..... ( -19.20) >DroSec_CAF1 8319 90 - 1 UGCCAAGGCAAUAAAUCGUUGCCUGCUGCUACGGCUGUUAAUUUGCAAUUCCGAUUUCAAUUUGAGCACAAUUUUUGGCUCUGAAAAAUA .((..(((((((.....)))))))...))..(((.(((......)))...))).(((((....((((.((.....))))))))))).... ( -21.50) >DroSim_CAF1 6433 90 - 1 UGCCAAGGCAAUAAAUCGUUGCCUGCUGCUACGGCUGUUAAUUUGCAAUUCCGAUUUCAAUUUGAGCACAAUUUUUGGCUCCGAAAAAUA .(((((((((((.....))))))((((((....))(((......))).................))))......)))))........... ( -21.40) >DroEre_CAF1 6494 90 - 1 UGCCAGGGCAAUAAAUCGUUGCCCGAUGCUACGGCUGUUAAUUUGCAAUUCCGAUUUCAAUUUGAGCACAGUUUUUGGCUCCGAAAAAUA .(((((((((((.....)))))))(.(((..(((.(((......)))...)))...((.....))))))......))))........... ( -22.60) >DroYak_CAF1 9667 90 - 1 UUCCAAGGCAAUAAAUCGUUGCCUGCUGCUACGGCUGUUAAUUUGCAAUUCCUAUUUCAAUUUGAGCACAAUUUUUGGCUCCGAAAAAUU .....(((((((.....)))))))(((((....))(((......))).................)))....((((((....))))))... ( -18.60) >consensus UGCCAAGGCAAUAAAUCGUUGCCUGCUGCUACGGCUGUUAAUUUGCAAUUCCGAUUUCAAUUUGAGCACAAUUUUUGGCUCCGAAAAAUA .(((((((((((.....)))))))(((((....))(((......))).................)))........))))........... (-18.36 = -18.64 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:03 2006