
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,768,780 – 1,768,876 |
| Length | 96 |
| Max. P | 0.992735 |

| Location | 1,768,780 – 1,768,876 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.37 |
| Mean single sequence MFE | -16.87 |
| Consensus MFE | -10.69 |
| Energy contribution | -10.80 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
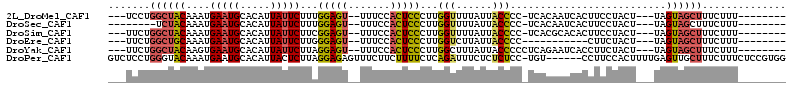
>2L_DroMel_CAF1 1768780 96 + 22407834 ---UCCUGGCUACAAAUGAAUGCACAUUAUUCUUUGGAGU--UUUCCACUCCCUUGGUUUUAUUACCCC-UCACAAUCACUUCCUACU---UAGUAGCUUUCUUU-------- ---....((((((....(((((.....)))))...(((((--.....)))))...(((......)))..-..................---..))))))......-------- ( -16.00) >DroSec_CAF1 25767 91 + 1 --------UCUACAAAUGAAUGCACAUUAUUCUUUGGAGU--UUUCCACUCCCUUGGUUUUAUUACCCC-UCACAAUCACUUCCUACU---UAGUAGCUUUCUUU-------- --------.........(((.((((..........(((((--.....)))))...(((......)))..-..................---..)).)).)))...-------- ( -11.60) >DroSim_CAF1 28091 96 + 1 ---UUCUGGCUACAAAUGAAUGCACAUUAUUCUUCGGAGU--UUUCCACUCCCUUGGUUUUAUUACCCC-UCACGCACACUUCCUACU---UAGUAGCUUUCUUU-------- ---....((((((....(((((.....)))))...(((((--.....)))))...(((......)))..-..................---..))))))......-------- ( -16.30) >DroEre_CAF1 25595 86 + 1 ---UUCUGGCUGCAAAUGAAUGCACAUUAUUCUUGGGAGU--UUUCCACUCCCUUGGUCUUAUUACCCC-----------CUUCUACU---UAGUAGCUUUCUUU-------- ---....((((((....(((((.....)))))..((((((--.....))))))..(((......)))..-----------........---..))))))......-------- ( -18.90) >DroYak_CAF1 34732 97 + 1 ---UUCUGGCUACAAGUGAAUGCACAUUAUUCUUAGGAGU--UUUCCACUCCCUUGGCUUUAUUACCCCCUCAGAAUCACCUUCUACU---UAGUAGCUUUCUUU-------- ---....(((((((((((((((.....)))))...(((((--.....)))))....................((((.....)))))))---).))))))......-------- ( -17.10) >DroPer_CAF1 27936 106 + 1 GUCUCCUGGGUACAAAUGAAUGCACAUUACUCUUAGGAGAGUUUCUUCUUUUCUCAGAUUUCUCUCUCC-UGU------CCUUCCACUUUUGAGUUGCUUUCUUUCUCCGUGG ....((((((...(((.(((.(((....((((.((((((((.....(((......))).....))))))-)).------............))))))).)))))).)))).)) ( -21.34) >consensus ___UUCUGGCUACAAAUGAAUGCACAUUAUUCUUAGGAGU__UUUCCACUCCCUUGGUUUUAUUACCCC_UCACAAUCACCUCCUACU___UAGUAGCUUUCUUU________ .......((((((....(((((.....)))))...(((((.......)))))...(((......)))..........................)))))).............. (-10.69 = -10.80 + 0.11)
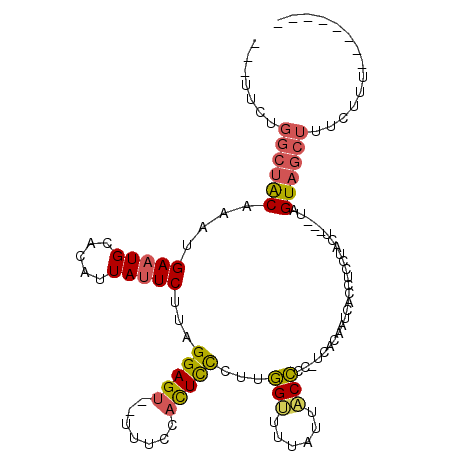
| Location | 1,768,780 – 1,768,876 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.37 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -11.61 |
| Energy contribution | -11.28 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1768780 96 - 22407834 --------AAAGAAAGCUACUA---AGUAGGAAGUGAUUGUGA-GGGGUAAUAAAACCAAGGGAGUGGAAA--ACUCCAAAGAAUAAUGUGCAUUCAUUUGUAGCCAGGA--- --------.......(((((..---......((((((.(((..-..(((......)))...(((((.....--)))))............))).))))))))))).....--- ( -21.20) >DroSec_CAF1 25767 91 - 1 --------AAAGAAAGCUACUA---AGUAGGAAGUGAUUGUGA-GGGGUAAUAAAACCAAGGGAGUGGAAA--ACUCCAAAGAAUAAUGUGCAUUCAUUUGUAGA-------- --------........((((..---.)))).((((((.(((..-..(((......)))...(((((.....--)))))............))).)))))).....-------- ( -18.40) >DroSim_CAF1 28091 96 - 1 --------AAAGAAAGCUACUA---AGUAGGAAGUGUGCGUGA-GGGGUAAUAAAACCAAGGGAGUGGAAA--ACUCCGAAGAAUAAUGUGCAUUCAUUUGUAGCCAGAA--- --------.......(((((.(---(((.((..(((..(((..-..(((......)))...(((((.....--)))))........)))..)))))))))))))).....--- ( -24.10) >DroEre_CAF1 25595 86 - 1 --------AAAGAAAGCUACUA---AGUAGAAG-----------GGGGUAAUAAGACCAAGGGAGUGGAAA--ACUCCCAAGAAUAAUGUGCAUUCAUUUGCAGCCAGAA--- --------........((((..---.))))..(-----------(.(((......)))..((((((.....--))))))..........((((......)))).))....--- ( -20.10) >DroYak_CAF1 34732 97 - 1 --------AAAGAAAGCUACUA---AGUAGAAGGUGAUUCUGAGGGGGUAAUAAAGCCAAGGGAGUGGAAA--ACUCCUAAGAAUAAUGUGCAUUCACUUGUAGCCAGAA--- --------.......(((((.(---(((.(((.((.(((((.....(((......)))..((((((.....--)))))).))))).....)).)))))))))))).....--- ( -25.60) >DroPer_CAF1 27936 106 - 1 CCACGGAGAAAGAAAGCAACUCAAAAGUGGAAGG------ACA-GGAGAGAGAAAUCUGAGAAAAGAAGAAACUCUCCUAAGAGUAAUGUGCAUUCAUUUGUACCCAGGAGAC ((((.(((...........)))....))))..((------..(-((((((.....(((......))).....))))))).........(((((......)))))))....... ( -25.10) >consensus ________AAAGAAAGCUACUA___AGUAGAAAGUGAUUGUGA_GGGGUAAUAAAACCAAGGGAGUGGAAA__ACUCCAAAGAAUAAUGUGCAUUCAUUUGUAGCCAGAA___ ................(((((....)))))..............(((((.......(((......))).....)))))...........((((......)))).......... (-11.61 = -11.28 + -0.33)

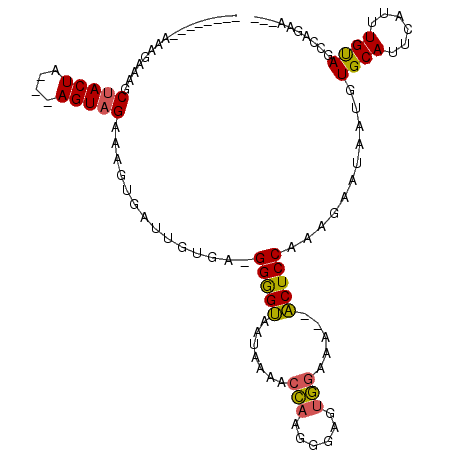
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:05 2006