
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,379,889 – 16,380,159 |
| Length | 270 |
| Max. P | 0.999882 |

| Location | 16,379,889 – 16,379,999 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.43 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -11.92 |
| Energy contribution | -12.90 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16379889 110 - 22407834 UUCUUAUUUAACCAAUCCCGGCUGGAAAGUUCUGAAUUUUCUUAAGCCGGC----------UUUAUUCGGAUUUUCAUAUUAUUUUACUCACAUCGAAUGCUAAAUUCAGUGUUUAAUUU ...........(((((.(((((((((((((.....)))))))..)))))).----------...))).))....................((((.((((.....)))).))))....... ( -19.60) >DroSec_CAF1 3573 101 - 1 UUCUUAUUUAACCAAUCCGGGCUGGAAAGUUCAGAGUUUUCUUAAGCCGGAGUUUUCUUUUCUUAUUAGGAUUUUCGAAU-------------------GCUAAAUUCAGUGUUAAAUAU ....(((((((((((((((((((....)))))((((..((((......))))..))))..........)))))...((((-------------------.....)))).).)))))))). ( -22.90) >DroYak_CAF1 3934 98 - 1 UUCUCAUUUAACCAAUCCCGGCUGGAAAGUUCCGAGUUUUCUUGAGCCGGC----------UUUAUUUGGAUUUUGCUAUUAUUGUACCCACACCGAAUGU--------GCAUUUG---- ...........((((..(((((((((....)))(((....))).)))))).----------.....))))....(((((((..(((....)))...)))).--------)))....---- ( -19.50) >consensus UUCUUAUUUAACCAAUCCCGGCUGGAAAGUUCAGAGUUUUCUUAAGCCGGC__________UUUAUUAGGAUUUUCAUAUUAUU_UAC_CACA_CGAAUGCUAAAUUCAGUGUUUAAU_U .................(((((((((((((.....)))))))..)))))).............................................(((((((......)))))))..... (-11.92 = -12.90 + 0.98)

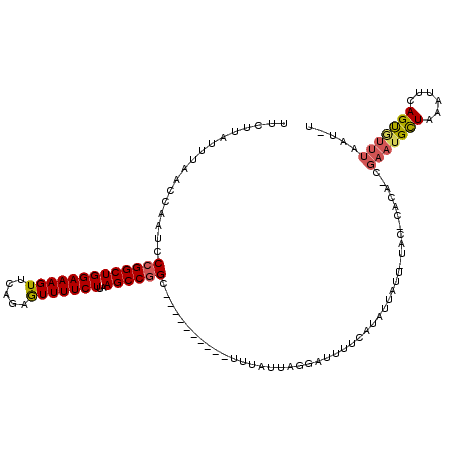

| Location | 16,379,999 – 16,380,119 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -29.38 |
| Energy contribution | -29.54 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.58 |
| SVM RNA-class probability | 0.999410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16379999 120 - 22407834 CAUUCCCUGUCGACACUUGUCACAUUCACAAAUGAAUGAAGUGCCAAAAGCACAACUGAGACGGGAAAACUCACGAAAAAUUAUGAUGCCAUGCCGUGACACAAGGUGUAACAGUAUACC ..(((((.((((((....))).((((((....))))))..((((.....))))......))))))))...(((((......((((....)))).))))).....((((((....)))))) ( -31.10) >DroSec_CAF1 3674 120 - 1 CAUUCCCCGUCGACACUUGUCACAUUCACAAAUGAAUGAAGUGCCAAAAGCACAACUGAGACGGGAAAACUCACGAAAAAUUAUGAUGCCAUGGCGUGACACAAGGUGUAACAGUAUACC .....(((((((((....))).((((((....))))))..((((.....))))......)))))).....(((((.....(((((....)))))))))).....((((((....)))))) ( -33.80) >DroSim_CAF1 1697 120 - 1 CAUUCCCCGUCGACACUUGUCACAUUCACAAAUGAAUGAAGUGCCAAAAGCACAACUGAGACGGGAAAACUCACGAAAAAUUAUGAUGCCAUGGCGUGACACAAGGUGUAACAGUAUACC .....(((((((((....))).((((((....))))))..((((.....))))......)))))).....(((((.....(((((....)))))))))).....((((((....)))))) ( -33.80) >DroEre_CAF1 1931 120 - 1 CAUUCCCCGUCGGCACUUGUCACAUUCACAAAUGAAUGAAGUGCCAAAAGCACAACUGAGACGGGAAAACUCUCGGAAAAUUAUGAUGGCUUGGCGUGACACAAGGUGUAACAGUGUACC (((.((((((((((((((....((((((....)))))))))))))..........((((((.(......)))))))........)))))...)).)))((((...........))))... ( -33.50) >DroYak_CAF1 4032 120 - 1 CAUUCCCUGUCGGCACUUGUCACAUUCACAAAUGAAUGAAGUGCCAAAAGCACAACUGAGACGGGAAAGCUCACGAAAAAUUAUGAUGCCAUGGCGUAACACAAGGUGUAACAGUAUACC ..(((((.((((((((((....((((((....)))))))))))))...((.....))..))))))))..................((((....)))).......((((((....)))))) ( -32.30) >consensus CAUUCCCCGUCGACACUUGUCACAUUCACAAAUGAAUGAAGUGCCAAAAGCACAACUGAGACGGGAAAACUCACGAAAAAUUAUGAUGCCAUGGCGUGACACAAGGUGUAACAGUAUACC .....(((((((((....))).((((((....))))))..((((.....))))......)))))).....(((((.....(((((....)))))))))).....((((((....)))))) (-29.38 = -29.54 + 0.16)

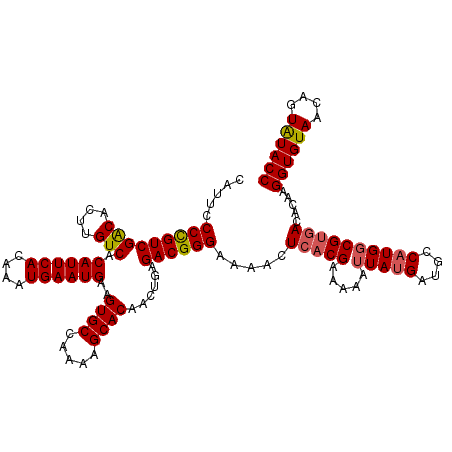
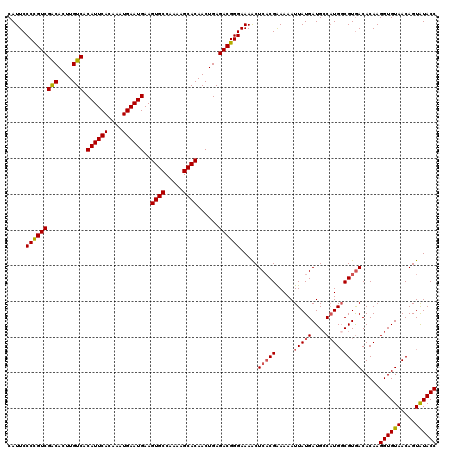
| Location | 16,380,039 – 16,380,159 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -35.36 |
| Energy contribution | -34.96 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.85 |
| SVM RNA-class probability | 0.999661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16380039 120 + 22407834 UUUUUCGUGAGUUUUCCCGUCUCAGUUGUGCUUUUGGCACUUCAUUCAUUUGUGAAUGUGACAAGUGUCGACAGGGAAUGAGAAUUGUUGCUGACGACGCUCAACUUUAUGUUGAUUACC ....((((.(((.......(((((....(.((.(((((((((((((((....))))))....))))))))).)).)..)))))......))).))))...(((((.....)))))..... ( -35.22) >DroSec_CAF1 3714 120 + 1 UUUUUCGUGAGUUUUCCCGUCUCAGUUGUGCUUUUGGCACUUCAUUCAUUUGUGAAUGUGACAAGUGUCGACGGGGAAUGAGAAUUGUUGCUGACGACGCUCAACUUUAUGUUGAUUACC ....((((.((((((((((((......((((.....))))..((((((....)))))).(((....))))))))))))...........))).))))...(((((.....)))))..... ( -36.10) >DroSim_CAF1 1737 120 + 1 UUUUUCGUGAGUUUUCCCGUCUCAGUUGUGCUUUUGGCACUUCAUUCAUUUGUGAAUGUGACAAGUGUCGACGGGGAAUGAGAAUUGUUGCUGACGACGCUCAACUUUAUGUUGAUUACC ....((((.((((((((((((......((((.....))))..((((((....)))))).(((....))))))))))))...........))).))))...(((((.....)))))..... ( -36.10) >DroEre_CAF1 1971 120 + 1 UUUUCCGAGAGUUUUCCCGUCUCAGUUGUGCUUUUGGCACUUCAUUCAUUUGUGAAUGUGACAAGUGCCGACGGGGAAUGAGAAUUGUUGCUGACGACGCUCAACUUUAUGUUGAUUACC .......((((((((((((((..((.....))...(((((((((((((....))))))....))))))))))))))))((((..(((((...)))))..)))))))))............ ( -37.10) >DroYak_CAF1 4072 120 + 1 UUUUUCGUGAGCUUUCCCGUCUCAGUUGUGCUUUUGGCACUUCAUUCAUUUGUGAAUGUGACAAGUGCCGACAGGGAAUGGGAAUUGUUGCUGACGACGCUCAACUUUAUGUUGAUUACC ....((((.(((.((((((((((.((((.((.((((.((((((((......))))).))).)))).)))))).))).))))))).....))).))))...(((((.....)))))..... ( -42.50) >consensus UUUUUCGUGAGUUUUCCCGUCUCAGUUGUGCUUUUGGCACUUCAUUCAUUUGUGAAUGUGACAAGUGUCGACGGGGAAUGAGAAUUGUUGCUGACGACGCUCAACUUUAUGUUGAUUACC ....((((.(((.......(((((....(.((.(((((((((((((((....))))))....))))))))).)).)..)))))......))).))))...(((((.....)))))..... (-35.36 = -34.96 + -0.40)


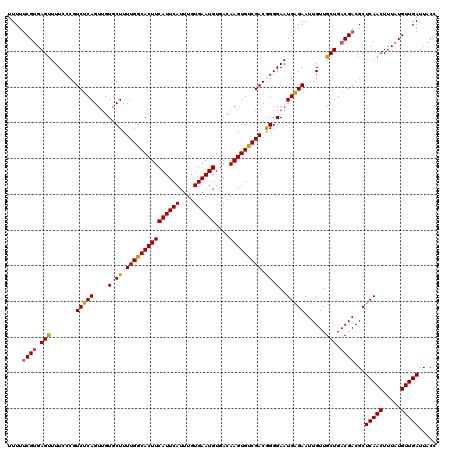
| Location | 16,380,039 – 16,380,159 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -29.46 |
| Energy contribution | -28.98 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.37 |
| SVM RNA-class probability | 0.999882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16380039 120 - 22407834 GGUAAUCAACAUAAAGUUGAGCGUCGUCAGCAACAAUUCUCAUUCCCUGUCGACACUUGUCACAUUCACAAAUGAAUGAAGUGCCAAAAGCACAACUGAGACGGGAAAACUCACGAAAAA .....(((((.....)))))..((((.(((................))).)))).((((((.((((((....))))))..((((.....))))......))))))............... ( -28.89) >DroSec_CAF1 3714 120 - 1 GGUAAUCAACAUAAAGUUGAGCGUCGUCAGCAACAAUUCUCAUUCCCCGUCGACACUUGUCACAUUCACAAAUGAAUGAAGUGCCAAAAGCACAACUGAGACGGGAAAACUCACGAAAAA .....(((((.....)))))((.......))..............(((((((((....))).((((((....))))))..((((.....))))......))))))............... ( -30.10) >DroSim_CAF1 1737 120 - 1 GGUAAUCAACAUAAAGUUGAGCGUCGUCAGCAACAAUUCUCAUUCCCCGUCGACACUUGUCACAUUCACAAAUGAAUGAAGUGCCAAAAGCACAACUGAGACGGGAAAACUCACGAAAAA .....(((((.....)))))((.......))..............(((((((((....))).((((((....))))))..((((.....))))......))))))............... ( -30.10) >DroEre_CAF1 1971 120 - 1 GGUAAUCAACAUAAAGUUGAGCGUCGUCAGCAACAAUUCUCAUUCCCCGUCGGCACUUGUCACAUUCACAAAUGAAUGAAGUGCCAAAAGCACAACUGAGACGGGAAAACUCUCGGAAAA .....(((((.....)))))((.......))..............(((((((((((((....((((((....)))))))))))))...((.....))..))))))............... ( -31.10) >DroYak_CAF1 4072 120 - 1 GGUAAUCAACAUAAAGUUGAGCGUCGUCAGCAACAAUUCCCAUUCCCUGUCGGCACUUGUCACAUUCACAAAUGAAUGAAGUGCCAAAAGCACAACUGAGACGGGAAAGCUCACGAAAAA .....(((((.....)))))...((((.(((...........(((((.((((((((((....((((((....)))))))))))))...((.....))..)))))))).))).)))).... ( -32.50) >consensus GGUAAUCAACAUAAAGUUGAGCGUCGUCAGCAACAAUUCUCAUUCCCCGUCGACACUUGUCACAUUCACAAAUGAAUGAAGUGCCAAAAGCACAACUGAGACGGGAAAACUCACGAAAAA .....(((((.....)))))((.......))..............(((((((((....))).((((((....))))))..((((.....))))......))))))............... (-29.46 = -28.98 + -0.48)

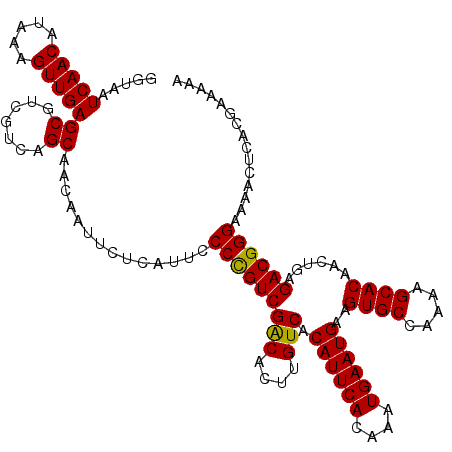
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:56 2006