
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,378,365 – 16,378,634 |
| Length | 269 |
| Max. P | 0.923095 |

| Location | 16,378,365 – 16,378,474 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.20 |
| Mean single sequence MFE | -25.49 |
| Consensus MFE | -17.56 |
| Energy contribution | -19.00 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16378365 109 - 22407834 AGUGCCAACUGCCAAAUGGCCAAGGCACCAUCCGCACCCACCAUCCGCCCACCUUUCGCCCACCUUACGGCCACCUUUCGCC-----------CACCACUCCCCUUCUGGCCACGUGUGC .(((((....(((....)))...)))))....(((((.........((.........)).........(((((.........-----------..............)))))..))))). ( -23.50) >DroSec_CAF1 2034 109 - 1 AGUGCCAACUGCCAAAUGGCCAAGGCACCACCCGCACCCACCAUCCGCCCACCUUUCGUCCACCUUACAGCCACCUUUCGCC-----------CACCACCCCCCUUCUGGCCACGUGGGC .(((((....(((....)))...)))))..................((((((.....((.......)).((((.........-----------..............))))...)))))) ( -24.20) >DroSim_CAF1 75 120 - 1 AGUGCCAACUGCCAAAUGGCCAAGGCACCACCCGCACCCACCAUCCGCCCACCUUUCGUCCACCUUUCGACCACCUUUCGACCACCUUUCGUACACCACCCCCCUUCUGGCCACGUGGGC .(((((....(((....)))...)))))..................((((((..............((((.......)))).....................((....))....)))))) ( -25.40) >DroEre_CAF1 216 113 - 1 AGUGCCAACUGCCAAAUGGCCAAGGCAGC------GCCCACCUUUCGGCCAGCUUUCCCCCACCUUUGGGCCAGCUUUCCCCCACCCUUCGCCCACCACCCUCUG-CUGACCACGUGGUA ..(((((...(((....)))...((((((------(((........))).((((..(((........)))..))))............................)-))).))...))))) ( -28.20) >DroYak_CAF1 2235 110 - 1 AGUGCCAACUGCCAAAUGGCCAAGGCACCAC---CACCCACCUUUCGACCACCAUCCACCCACCUUUCGGCCACCUUUCGCCCACC-------CACCAACCCCCGUUUGGCCACGUGGGA .(((((....(((....)))...)))))...---..(((((.....((......))............(((((.............-------..............)))))..))))). ( -26.13) >consensus AGUGCCAACUGCCAAAUGGCCAAGGCACCACCCGCACCCACCAUCCGCCCACCUUUCGCCCACCUUUCGGCCACCUUUCGCCCACC_______CACCACCCCCCUUCUGGCCACGUGGGC .(((((....(((....)))...))))).............................((((((.....(((((..................................)))))..)))))) (-17.56 = -19.00 + 1.44)

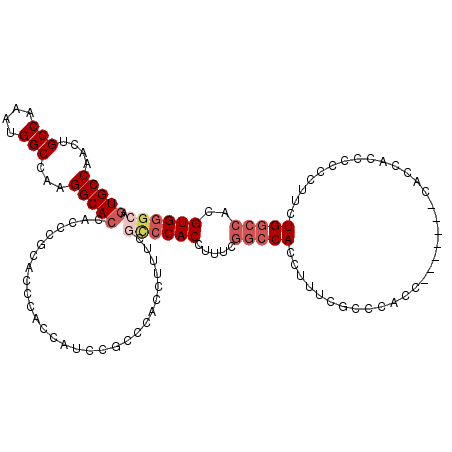

| Location | 16,378,474 – 16,378,594 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -23.68 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16378474 120 + 22407834 CGAAUGCAAUUUCAGUAAUUAAAAUAAACAACACACGAUGACGACGUGCAGUUUGACUGCACCGAUGGGCUUUUGACCAGAUCGACGACAACAUCGUCAUCAACAUAUCAUUAUCAGACA .(((......))).......................((((((((.((((((.....))))))((((.(((....).))..)))).........))))))))................... ( -28.00) >DroSec_CAF1 2143 120 + 1 CGAAUGCAAUUUCAGUAAUUAAAAUAAACAACACACGAUGACGACGUGCAGUUUGACUGCACCGAUGGGCUUUUGACCAGAUCGACGACAACAUCGUCAUCAACAUAUCAUUAUCAGACA .(((......))).......................((((((((.((((((.....))))))((((.(((....).))..)))).........))))))))................... ( -28.00) >DroSim_CAF1 195 120 + 1 CGAAUGCAAUUUCAGUAAUUAAAAUAAACAACACACGAUGACGACGUGCAGUUUGACUGCACCGAUGGGCUUUUGACCAGAUCGACGACAACAUCGUCAUCAACAUAUCAUUAUCAGACA .(((......))).......................((((((((.((((((.....))))))((((.(((....).))..)))).........))))))))................... ( -28.00) >DroEre_CAF1 329 117 + 1 CGAAUGCAAUUUCAGUAAUUAAAAUAAACAACACACGACGACGACGUGCAGUUUGACUGCAGCGACGGGCUUUUGACCAGAUCG---ACAACAUCGUCAUCAACAUAUCAUUAUCAGACA .(((......))).......................((.(((((.((((((.....)))).((((..(((....).))...)))---.).)).))))).))................... ( -20.00) >DroYak_CAF1 2345 120 + 1 CGAAUGCAAUUUCAGUAAUUAAAAUAAACAACACACGAUGACGAAGUGGAGUUUGACUGCACCGAUGGGCUUUUGACCAGAUCGACGACAACAUCGUCAUCAACAUAUCAUUAUCAGACA .(((......))).......................((((((((.(((.((.....)).)))((((.(((....).))..)))).........))))))))................... ( -23.70) >consensus CGAAUGCAAUUUCAGUAAUUAAAAUAAACAACACACGAUGACGACGUGCAGUUUGACUGCACCGAUGGGCUUUUGACCAGAUCGACGACAACAUCGUCAUCAACAUAUCAUUAUCAGACA .(((......))).......................((((((((.((((((.....))))))((((((........)))..))).........))))))))................... (-23.68 = -24.48 + 0.80)



| Location | 16,378,514 – 16,378,634 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -30.84 |
| Consensus MFE | -25.14 |
| Energy contribution | -26.74 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16378514 120 + 22407834 ACGACGUGCAGUUUGACUGCACCGAUGGGCUUUUGACCAGAUCGACGACAACAUCGUCAUCAACAUAUCAUUAUCAGACAAUCCUGGCAUCCAUUUGGGCCAAUUGCUUUGUCAAAAUAA ..(((((((((.....))))))....((((..(((.((((((.(((((.....)))))...............((((......)))).....))))))..)))..)))).)))....... ( -32.80) >DroSec_CAF1 2183 120 + 1 ACGACGUGCAGUUUGACUGCACCGAUGGGCUUUUGACCAGAUCGACGACAACAUCGUCAUCAACAUAUCAUUAUCAGACAAUCCUGGCAUCCAUUUGGGCCAAUUGCUUUGUCAAAAUAA ..(((((((((.....))))))....((((..(((.((((((.(((((.....)))))...............((((......)))).....))))))..)))..)))).)))....... ( -32.80) >DroSim_CAF1 235 120 + 1 ACGACGUGCAGUUUGACUGCACCGAUGGGCUUUUGACCAGAUCGACGACAACAUCGUCAUCAACAUAUCAUUAUCAGACAAUCCUGGCAUCCAUUUGGGCCAAUUGCUUUGUCAAAAUAA ..(((((((((.....))))))....((((..(((.((((((.(((((.....)))))...............((((......)))).....))))))..)))..)))).)))....... ( -32.80) >DroEre_CAF1 369 117 + 1 ACGACGUGCAGUUUGACUGCAGCGACGGGCUUUUGACCAGAUCG---ACAACAUCGUCAUCAACAUAUCAUUAUCAGACAAUCCUUGCAUCCAUUUGGGCCAAUUGCUUUGUCAAAAUAA ..(((((((((.....)))))((((..(((.........(((.(---((......)))))).............................((....)))))..))))..))))....... ( -25.50) >DroYak_CAF1 2385 120 + 1 ACGAAGUGGAGUUUGACUGCACCGAUGGGCUUUUGACCAGAUCGACGACAACAUCGUCAUCAACAUAUCAUUAUCAGACAAUCCUUGCGUCCAUUUGGGCCAAUUGCUUUGUCAAAAUAA ((((((((((((((((.........(((........)))(((.(((((.....))))))))............))))))..)))(((.((((....)))))))..)))))))........ ( -30.30) >consensus ACGACGUGCAGUUUGACUGCACCGAUGGGCUUUUGACCAGAUCGACGACAACAUCGUCAUCAACAUAUCAUUAUCAGACAAUCCUGGCAUCCAUUUGGGCCAAUUGCUUUGUCAAAAUAA ..(((((((((.....))))))....((((..(((.((((((.(((((.....)))))...............((((......)))).....))))))..)))..)))).)))....... (-25.14 = -26.74 + 1.60)


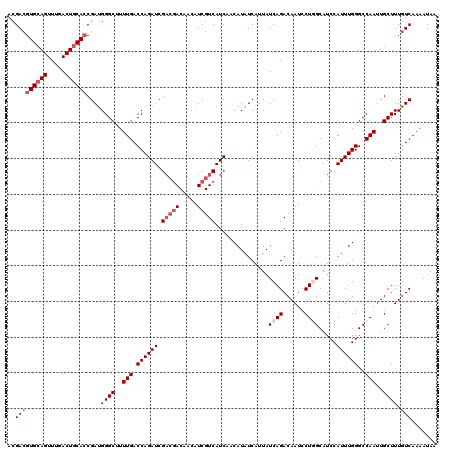
| Location | 16,378,514 – 16,378,634 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -31.38 |
| Energy contribution | -32.86 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16378514 120 - 22407834 UUAUUUUGACAAAGCAAUUGGCCCAAAUGGAUGCCAGGAUUGUCUGAUAAUGAUAUGUUGAUGACGAUGUUGUCGUCGAUCUGGUCAAAAGCCCAUCGGUGCAGUCAAACUGCACGUCGU .......(((...((..((((((...((.((((.(((.((((((...((((.....))))..)))))).))).)))).))..))))))..))......((((((.....))))))))).. ( -37.80) >DroSec_CAF1 2183 120 - 1 UUAUUUUGACAAAGCAAUUGGCCCAAAUGGAUGCCAGGAUUGUCUGAUAAUGAUAUGUUGAUGACGAUGUUGUCGUCGAUCUGGUCAAAAGCCCAUCGGUGCAGUCAAACUGCACGUCGU .......(((...((..((((((...((.((((.(((.((((((...((((.....))))..)))))).))).)))).))..))))))..))......((((((.....))))))))).. ( -37.80) >DroSim_CAF1 235 120 - 1 UUAUUUUGACAAAGCAAUUGGCCCAAAUGGAUGCCAGGAUUGUCUGAUAAUGAUAUGUUGAUGACGAUGUUGUCGUCGAUCUGGUCAAAAGCCCAUCGGUGCAGUCAAACUGCACGUCGU .......(((...((..((((((...((.((((.(((.((((((...((((.....))))..)))))).))).)))).))..))))))..))......((((((.....))))))))).. ( -37.80) >DroEre_CAF1 369 117 - 1 UUAUUUUGACAAAGCAAUUGGCCCAAAUGGAUGCAAGGAUUGUCUGAUAAUGAUAUGUUGAUGACGAUGUUGU---CGAUCUGGUCAAAAGCCCGUCGCUGCAGUCAAACUGCACGUCGU ...(((((((...((((((.((((....))..))...))))))..........((.(((((..((...))..)---)))).)))))))))...((.((.(((((.....))))))).)). ( -28.90) >DroYak_CAF1 2385 120 - 1 UUAUUUUGACAAAGCAAUUGGCCCAAAUGGACGCAAGGAUUGUCUGAUAAUGAUAUGUUGAUGACGAUGUUGUCGUCGAUCUGGUCAAAAGCCCAUCGGUGCAGUCAAACUCCACUUCGU ....((((((.......((((((...((.(((((((.(.(((((...((((.....))))..)))))).))).)))).))..))))))..((((...)).)).))))))........... ( -29.40) >consensus UUAUUUUGACAAAGCAAUUGGCCCAAAUGGAUGCCAGGAUUGUCUGAUAAUGAUAUGUUGAUGACGAUGUUGUCGUCGAUCUGGUCAAAAGCCCAUCGGUGCAGUCAAACUGCACGUCGU .......(((...((..((((((...((.((((.(((.((((((...((((.....))))..)))))).))).)))).))..))))))..))......((((((.....))))))))).. (-31.38 = -32.86 + 1.48)


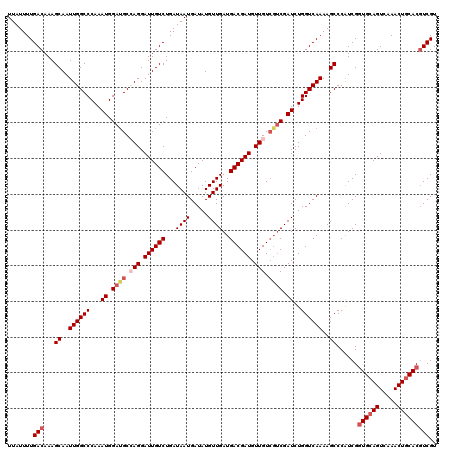
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:51 2006