
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,768,157 – 1,768,254 |
| Length | 97 |
| Max. P | 0.685996 |

| Location | 1,768,157 – 1,768,254 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.64 |
| Mean single sequence MFE | -18.48 |
| Consensus MFE | -13.01 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1768157 97 + 22407834 UAUAGUUUUGCACUUAGCAAUUAACGAGUUUACUUU-UUUUAGUGCUACUCCCAGUGCCCGCAUCAAGUGGACAUAUCCUCCUCCACC-----------UCUCCACU-----------CC ....((...((((((((((...((.(((....))).-))....))))).....)))))..))....((((((................-----------..))))))-----------.. ( -16.77) >DroPse_CAF1 28354 118 + 1 UAUGGUUUUGCACUUAGCAAUUAACGAGUUUACUUU--UUUAGUGCUGCUCCCAGAACCCGCAUGAAGUGGACAUCUCCUCCUCUUCCAUCCUCAUCCUCAUCCACACCCAUCCCCGUCC ...(((((((....(((((...((.(((....))).--))...)))))....))))))).....((((.(((.......))).))))................................. ( -20.60) >DroSec_CAF1 25185 97 + 1 UAUGGUUUUGCACUUAGCAAUUAACGAGUUUACUUU-UUUUAGUGCUACUCCCAGUGCCCGCAUGAAGUGACCAUAUCCUCCUCCACU-----------CCUCCACA-----------GU ((((((...((((((((((...((.(((....))).-))....))))).....))))).(((.....)))))))))............-----------........-----------.. ( -17.80) >DroSim_CAF1 27469 98 + 1 UAUGGUUUUGCACUUAGCAAUUAACGAGUUUACUUUUUUUUAGUGCUACUCCCAGUGCCCGCAUCAAGUGGACAUAUCCUCCUCCACU-----------CCUCCACU-----------GC ..(((....((((((((((...((.(((....))).)).....))))).....)))))........((((((..........))))))-----------...)))..-----------.. ( -18.00) >DroEre_CAF1 24955 94 + 1 UAUGCUUUUGCACUUAGCAAUUAACGAGUUUACUUU-UUUUAGUGCUACUCCCAGUGCCCGCAUCAAGUGGACAUAUCCUCCUCCACU-----------UUUCCAC-------------- .((((....((((((((((...((.(((....))).-))....))))).....)))))..)))).(((((((..........))))))-----------)......-------------- ( -20.30) >DroYak_CAF1 34126 86 + 1 UAUGGUUUUGCACUUAGCAAUUAACGAGUUUACUUU-UUUUAGUGCUACUCCCAGUGCCCGCAUCAAGUGGACAUAUCCUCCUCCAC--------------------------------- ((((.....((((((((((...((.(((....))).-))....))))).....)))))((((.....)))).))))...........--------------------------------- ( -17.40) >consensus UAUGGUUUUGCACUUAGCAAUUAACGAGUUUACUUU_UUUUAGUGCUACUCCCAGUGCCCGCAUCAAGUGGACAUAUCCUCCUCCACU___________CCUCCAC_____________C ..(((..((((.....)))).....((((.((((.......))))..)))))))(((.((((.....)))).)))............................................. (-13.01 = -13.70 + 0.69)

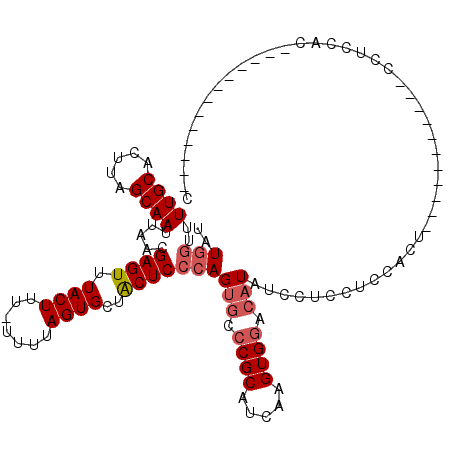
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:03 2006