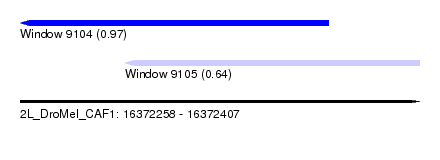
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,372,258 – 16,372,407 |
| Length | 149 |
| Max. P | 0.973107 |

| Location | 16,372,258 – 16,372,373 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -24.21 |
| Energy contribution | -24.65 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.973107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16372258 115 - 22407834 UCUACAUGCCGCGUGGCUCUAAUGGGCCAACUUGUAAGCCUCCACUGCUGCCCCGAGGGCAGUACUUCCCGCGA-CAAUCGCGC-CCCAAUACUCUGGGAAUCCCCAAACCUGAAAU .........((((((((((....))))))..((((..((......((((((((...))))))))......)).)-))).)))).-((((......)))).................. ( -34.40) >DroSim_CAF1 47531 115 - 1 UCUACAAGGCGCGUGGCUCUAAUGGGCCAACUUGUAAGCCUCCACUGCUGCCCCGAGGGCAGUACUUCCCGCGA-CAAUCCCGC-CCCAAUACUCUGGGAAUCCCCAAACCUGAAAU .......((.(((((((((....))))))..((((..((......((((((((...))))))))......)).)-)))...)))-.)).......((((....)))).......... ( -36.00) >DroEre_CAF1 47781 87 - 1 UCCACCAGGCGCGUGGCCCUAAUGGGCCAACUUGUAAGCCUGCACUGCUGCC-CGAGGGCAGUA----------------------------CUCUCGGAAUCCACG-ACAUGGAAU (((..((((((((((((((....))))))...)))..)))))...(((((((-....)))))))----------------------------.....))).((((..-...)))).. ( -37.90) >DroYak_CAF1 47946 116 - 1 UCUACAAGGCGCGUGGCUCUAAUGGGCCAACUUGUAAGCCUCCAAUGCUGCCCCGAGGGCAGUACUUCCCGCGUCCAAUACCGCGCCCAGUACUCUCCGAAUUCCCA-ACGUGGAAU ..(((..((((((((((((....))))))..(((...((......((((((((...))))))))......))...)))...))))))..)))........(((((..-....))))) ( -40.10) >consensus UCUACAAGGCGCGUGGCUCUAAUGGGCCAACUUGUAAGCCUCCACUGCUGCCCCGAGGGCAGUACUUCCCGCGA_CAAUCCCGC_CCCAAUACUCUCGGAAUCCCCA_ACCUGAAAU ((((((((.....((((((....)))))).)))))..........(((((((.....))))))).................................)))................. (-24.21 = -24.65 + 0.44)

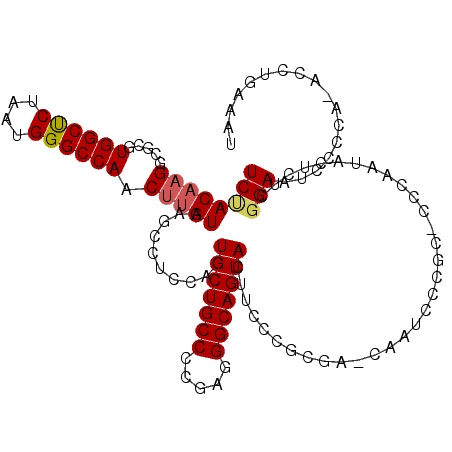

| Location | 16,372,297 – 16,372,407 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -31.41 |
| Consensus MFE | -18.74 |
| Energy contribution | -20.10 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16372297 110 - 22407834 CCUUGGCCCCUCCCAAAAUCCACAAA------AGAGCGACUCUACAUGCCGCGUGGCUCUAAUGGGCCAACUUGUAAGCCUCCACUGCUGCCCCGAGGGCAGUACU-UCCCGCGA-CA ..(((((((.................------(((((.((.(........).)).)))))...))))))).......((......((((((((...))))))))..-....))..-.. ( -32.35) >DroPse_CAF1 41511 91 - 1 -------------CACCGCCCACAAA------AGA--CACUCU--CAGACACG-GCCUCUAAUGGGCCAACUUGUAAGCCUCCAAUGCUGCCCCGAGGGCAGUACUCUCCCGCGU--- -------------...(((.......------.((--..((..--(((....(-((((.....)))))...)))..))..))...((((((((...)))))))).......))).--- ( -24.50) >DroSim_CAF1 47570 110 - 1 CCUUGGCCCCUCCCAAAAUCCACAAA------AGAGCGACUCUACAAGGCGCGUGGCUCUAAUGGGCCAACUUGUAAGCCUCCACUGCUGCCCCGAGGGCAGUACU-UCCCGCGA-CA ..((((......))))..........------...(((....((((((.....((((((....)))))).)))))).........((((((((...))))))))..-...)))..-.. ( -33.90) >DroEre_CAF1 47805 97 - 1 CAUUGGCCCCACCCAAAGUCCACAAA------AGAGCGACUCCACCAGGCGCGUGGCCCUAAUGGGCCAACUUGUAAGCCUGCACUGCUGCC-CGAGGGCAGUA-------------- ..((((......))))(((((.....------...).))))....((((((((((((((....))))))...)))..)))))...(((((((-....)))))))-------------- ( -35.30) >DroYak_CAF1 47985 117 - 1 CCUUGGCCCCUCCCAAAGUCCACAAGCGACACAGAGCGACUCUACAAGGCGCGUGGCUCUAAUGGGCCAACUUGUAAGCCUCCAAUGCUGCCCCGAGGGCAGUACU-UCCCGCGUCCA ..(((((((........((((....).)))..(((((.((.(........).)).)))))...))))))).......((......((((((((...))))))))..-....))..... ( -37.90) >DroPer_CAF1 42893 91 - 1 -------------CACCGCCCACAAA------AGA--CACUCU--CAGACACG-GCCUCUAAUGGGCCAACUUGUAAGCCUCCAAUGCUGCCCCGAGGGCAGUACUCUCCCGCGU--- -------------...(((.......------.((--..((..--(((....(-((((.....)))))...)))..))..))...((((((((...)))))))).......))).--- ( -24.50) >consensus CCUUGGCCCCUCCCAAAGUCCACAAA______AGAGCGACUCUACAAGGCGCGUGGCUCUAAUGGGCCAACUUGUAAGCCUCCAAUGCUGCCCCGAGGGCAGUACU_UCCCGCGU___ .....................((((..........(((.((.....)).)))..(((((....)))))...))))..........(((((((.....))))))).............. (-18.74 = -20.10 + 1.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:44 2006