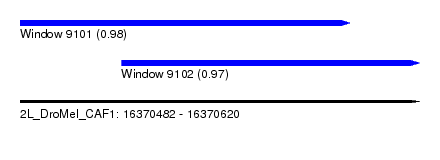
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,370,482 – 16,370,620 |
| Length | 138 |
| Max. P | 0.976017 |

| Location | 16,370,482 – 16,370,596 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -23.13 |
| Energy contribution | -22.82 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
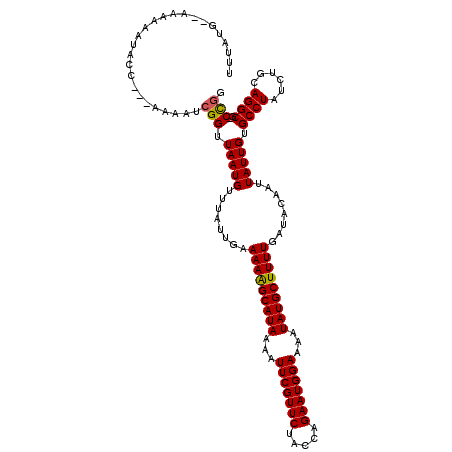
>2L_DroMel_CAF1 16370482 114 + 22407834 UAUAUG--AAAAAAUACC---AAAAUCGGUUAAUGUUUAUUGAAAAAGCAUAAAAUUCGUUCUACCAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCG-CCGG ...(((--((.....(((---......))).....)))))...(((((((((...(((((((.....)))))))...)))))))))............((((((......)))))-)... ( -26.80) >DroVir_CAF1 29851 114 + 1 UUUAUG--AAAAAAUACC---AAAAUCGGUUAAUGUUUAUUGAAAAGGCAUAAAAUUCGUUCUACUAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCG-CUGG ((.(((--((.....(((---......))).....))))).))(((((((((...(((((((.....)))))))...)))))))))............((((((......)))))-)... ( -27.00) >DroPse_CAF1 39987 116 + 1 UAUAUGAGAAAAAAUACC---AAAAUCGGUUAAUGUUUAUUGAAAAAGCAUAAAAUUCGUUCUACCAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCG-CCGG ..................---.....(((((((((........(((((((((...(((((((.....)))))))...)))))))))........))))).((((......)))))-))). ( -26.29) >DroGri_CAF1 60278 114 + 1 UUUAUG--AAAAAAUACC---AAAAUCGGUUAAUGUUUAUUGAAAAGGCAUAAAAUUCGUUCUACUAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCG-CCAG ((.(((--((.....(((---......))).....))))).))(((((((((...(((((((.....)))))))...)))))))))............((((((......)))))-)... ( -26.60) >DroWil_CAF1 50433 118 + 1 GUUAUG--AAAAAAUACCGAAAAAAUCGGUUAAUGUUUAUUGAAAAAGCAUAAAAUUCGUUCUACCAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCAACCGG ...(((--((.....(((((.....))))).....)))))...(((((((((...(((((((.....)))))))...))))))))).............(((((......)))))..... ( -28.10) >DroMoj_CAF1 46960 114 + 1 UUUAUG--AAAAAAUACC---AAAAUCGGUUAAUGUUUAUUGAAAAGGCAUAAAAUUCGUUCUACUAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCA-CUGC ((.(((--((.....(((---......))).....))))).))(((((((((...(((((((.....)))))))...)))))))))............((((((......)))))-)... ( -27.40) >consensus UUUAUG__AAAAAAUACC___AAAAUCGGUUAAUGUUUAUUGAAAAAGCAUAAAAUUCGUUCUACCAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCG_CCGG ..........................(((.(((((........(((((((((...(((((((.....)))))))...)))))))))........))))).((((......))))..))). (-23.13 = -22.82 + -0.31)

| Location | 16,370,517 – 16,370,620 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -25.13 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.05 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
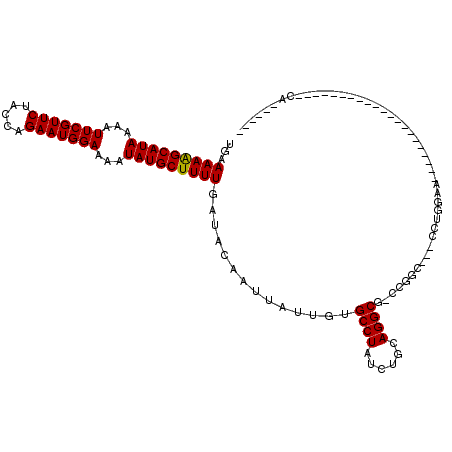
>2L_DroMel_CAF1 16370517 103 + 22407834 UGAAAAAGCAUAAAAUUCGUUCUACCAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCG-CCGGC---CCUG-AA-AACGCACCC-----------CAAUCCUA ...(((((((((...(((((((.....)))))))...)))))))))............((((.......((((.(-....)---))))-..-...))))..-----------........ ( -24.50) >DroVir_CAF1 29886 92 + 1 UGAAAAGGCAUAAAAUUCGUUCUACUAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCG-CUGGCAAACCUGGGA---------------------CA------ ...(((((((((...(((((((.....)))))))...)))))))))............((((((......)))))-)..............---------------------..------ ( -23.30) >DroGri_CAF1 60313 92 + 1 UGAAAAGGCAUAAAAUUCGUUCUACUAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCG-CCAGCAAACCUUGAA---------------------CC------ ...(((((((((...(((((((.....)))))))...)))))))))............((((((......)))))-)..............---------------------..------ ( -22.90) >DroWil_CAF1 50471 116 + 1 UGAAAAAGCAUAAAAUUCGUUCUACCAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCAACCGGC---GAUA-CAGUAUCCACGGCAUCCAUAGACCAUCCAUA ...(((((((((...(((((((.....)))))))...)))))))))(((((...((((((((((......)))).....))---))))-..)))))...(((.......).))....... ( -26.90) >DroMoj_CAF1 46995 92 + 1 UGAAAAGGCAUAAAAUUCGUUCUACUAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCA-CUGCAAAACCUGGCA---------------------CA------ ...(((((((((...(((((((.....)))))))...)))))))))...........((((((....(((((...-)))))......))))---------------------))------ ( -27.80) >DroPer_CAF1 41379 104 + 1 UGAAAAAGCAUAAAAUUCGUUCUACCAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCG-CCGGC---CCUGGAA-AACGCACCC-----------CAAACCUU ...(((((((((...(((((((.....)))))))...)))))))))............((((((......)))))-).((.---..(((..-........)-----------))..)).. ( -25.40) >consensus UGAAAAAGCAUAAAAUUCGUUCUACCAGAAUGGAAAAUAUGCUUUUGAUACAAUUAUUGUGCCUAUCUGCAGGCG_CCGGC___CCUGGAA_____________________CA______ ...(((((((((...(((((((.....)))))))...)))))))))..............((((......)))).............................................. (-20.30 = -20.05 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:41 2006