
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,367,709 – 16,367,816 |
| Length | 107 |
| Max. P | 0.589897 |

| Location | 16,367,709 – 16,367,816 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -18.02 |
| Energy contribution | -19.22 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16367709 107 - 22407834 CAAGAGAAGUGCUGCUUGCGGUCGUUUUAAACAAACCGCAGGUGUUGCAUACAUUUGCCAGCUUUGCAUUAAUUAUGACGUGUGUCUCGGGAG-AA----AGGCGUGGCAAA ........((((.(((((((((.((.....))..)))))))))...))))...((((((((((((...........(((....)))((....)-))----)))).))))))) ( -33.00) >DroVir_CAF1 26402 95 - 1 --UGGG----AAUUCUUGCGGUCUUUUUAAACAAACCGCAGGUGUUGCAUACAUUGGCCAGCUUUGCAUUAAUUAUGAUGUUUUGUGCAGCAUGGA-------ACUC----A --.(((----....((((((((............))))))))(((((((((....((....))..((((((....))))))..)))))))))....-------.)))----. ( -24.90) >DroPse_CAF1 37688 107 - 1 ---GGGAGGCAAUGCUUGUGGUCGUUUUAAACAAACCGCAGGUGUUGCAUACAUUUGCCAGCUUUGCAUUAAUUAUGAUGUGUGUCUCGGUGGGGGCGGCCGGAGGGGCA-- ---.(...((((((((((((((.((.....))..))))))))))))))...)...((((..((((((((((....)))))).((((((....))))))...)))).))))-- ( -36.00) >DroEre_CAF1 40845 108 - 1 CAUGAGAAGUGCUGCUUGCGGUCGUUUUAAACAAACCGCAGGUGUUGCAUACAUUUGCCAGCUUUGCAUUAAUUAUGACGUGUGUCUCGGUGGAAG----GGACGUGGAAGA (((((..(((((.(((((((((.((.....))..)))))))))((((((......)).))))...)))))..)))))......(((((.......)----))))........ ( -32.00) >DroMoj_CAF1 43756 99 - 1 --UGGG----AAUGCUUGCGGUCUUUUUAAACAAACCGCAGGUGUUGCAUACAUCGGCCAGCUUUGCAUUAAUUAUGAUGUUUUGUGCAGUGCGGA-------ACUGGCAGA --..(.----((((((((((((............)))))))))))).).....((.(((((.((((((((...((..(....)..)).))))))))-------.))))).)) ( -34.00) >DroPer_CAF1 37874 97 - 1 ---GGGGGGCAAUGCUUGUGGUCGUUUUAAACAAACCGCAGGUGUUGCAUACAUUUGCCAGCUUUGCAUUAAUUAUGAUGUGUGUCUCGGUGGGGGCGGC------------ ---.(...((((((((((((((.((.....))..))))))))))))))...)....(((......((((((....))))))..(((((....))))))))------------ ( -32.40) >consensus __UGGGA_G_AAUGCUUGCGGUCGUUUUAAACAAACCGCAGGUGUUGCAUACAUUUGCCAGCUUUGCAUUAAUUAUGAUGUGUGUCUCGGUGGGGA_______AGUGGCA_A ...((((.((((((((((((((............)))))))))))))).................((((((....))))))...))))........................ (-18.02 = -19.22 + 1.20)

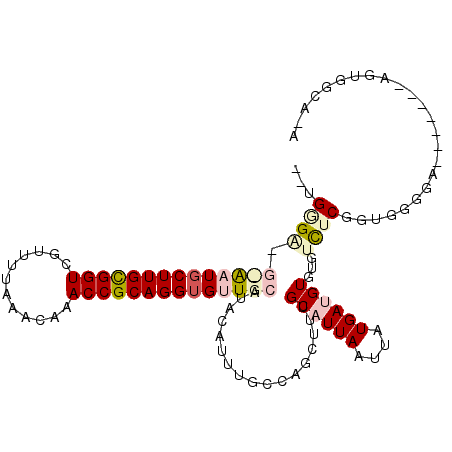
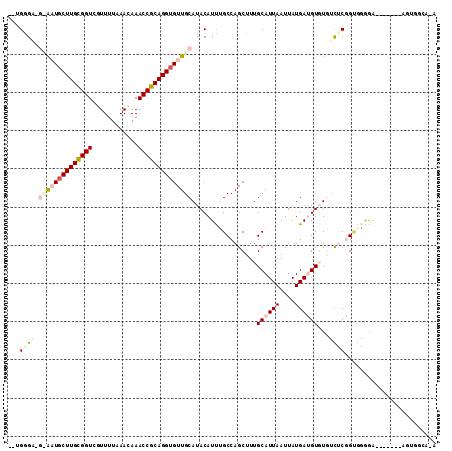
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:38 2006