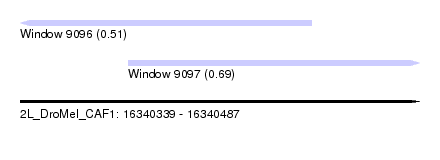
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,340,339 – 16,340,487 |
| Length | 148 |
| Max. P | 0.693451 |

| Location | 16,340,339 – 16,340,447 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -42.23 |
| Consensus MFE | -27.85 |
| Energy contribution | -28.10 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
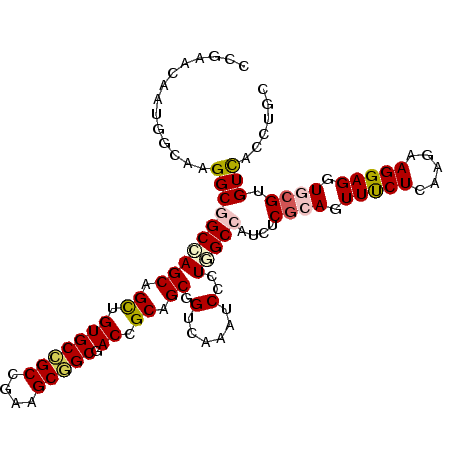
>2L_DroMel_CAF1 16340339 108 - 22407834 CCGAAUAGUGGAAAGGCGGCGAGCAGUUGUGCCGCCGAAGCGGCGACGGCUGCCGUGAAGUCAUUGGCCAUCUCGCAGUUCCUGAAGAAGGAGGUGCGUGUCACCUGC .......((((...(((.(((.(((((((((((((....))))).)))))))))))(....)....)))..(.((((.(((((.....))))).)))).))))).... ( -45.50) >DroVir_CAF1 1435 108 - 1 CCCAACAACGCCAUGGCCGCCAGCAGCUGUGCCGCCGAGGCGGCUACUGCUGCGGUCAAAUCGCUUGCCAUAUCGCAGUUUCUAAAAAAGGAGGUGCGUGUUACAUGU ......(((((..(((((((.(((((.((.(((((....)))))))))))))))))))................(((.(((((.....))))).))))))))...... ( -43.60) >DroPse_CAF1 2119 108 - 1 CCGAACAGUGGCAAGGCGGCCAGCAGCUGUGCCGCCGAAGCGGCGACAGCGGCUGUGAAAUCCCUGGCCAUCUCGCAGUUUCUCAAGAAGGAGGUUCGUGUCACCUGU .(((((.(((((.(((...(((((.((((((((((....))))).))))).)))).).....))).)))))..........(((......)))))))).......... ( -42.70) >DroWil_CAF1 2205 108 - 1 CCCAAUAAUGGAAAGGCGGCCAGCAGUUGUGCCGCUGAAGCGGCAACCGCAGCUGUCAAAUCUUUGGCCAUCUCUCAGUUUCUCAAGAAGGAGGUUCGUGUCACGUGC .(((....)))...(((((((((((((((((((((....)))))....))))))))........)))))((((((....(((....)))))))))....)))...... ( -36.00) >DroMoj_CAF1 19150 108 - 1 CCUAACAACGGCAUGGCCGCAAGCAGCUGUGCCGCAGAGGCAGCUACUGCAGCGGUCAAAUCGCUUGCCAUAUCGCAGUUUCUAAAAAAGGAGGUGCGUGUUACAUGC ..(((((..(((((((((((..((((..(((((.....)))).)..)))).))))))).......))))....((((.(((((.....))))).)))))))))..... ( -41.01) >DroAna_CAF1 12128 108 - 1 CCGAACAAUGGGAAGGCGGCCAGCAGUUGUGCGGCGGAAGCUGCCACCGCGGCGGUCAAAUCCCUGGCCAUUUCGCAGUUCCUCAAGAAGGAGGUGCGUGUCACCUGC .........((((.(((.(((.((.((.(.(((((....)))))))).))))).)))...))))((((.....((((.(((((.....))))).)))).))))..... ( -44.60) >consensus CCGAACAAUGGCAAGGCGGCCAGCAGCUGUGCCGCCGAAGCGGCGACCGCAGCGGUCAAAUCCCUGGCCAUCUCGCAGUUUCUCAAGAAGGAGGUGCGUGUCACCUGC ..............((((((((((.((.(((((((....))))).)).)).)).(......)..)))))....((((.(((((.....))))).)))).)))...... (-27.85 = -28.10 + 0.25)


| Location | 16,340,379 – 16,340,487 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.49 |
| Mean single sequence MFE | -43.47 |
| Consensus MFE | -27.86 |
| Energy contribution | -30.82 |
| Covariance contribution | 2.95 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
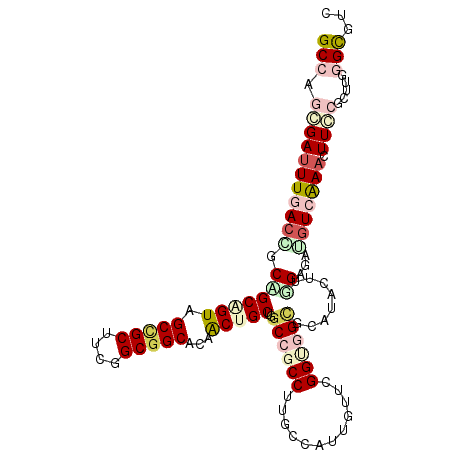
>2L_DroMel_CAF1 16340379 108 + 22407834 GCCAAUGACUUCACGGCAGCCGUCGCCGCUUCGGCGGCACAACUGCUCGCCGCCUUUCCACUAUUCGGUGGCGCAUACUUGGAGAUGUCAAACUUCCGCUUCGGCGUC .....((((.((..(((((..((.(((((....)))))))..)))))(((((((............)))))))..........)).))))......(((....))).. ( -43.60) >DroVir_CAF1 1475 108 + 1 GCAAGCGAUUUGACCGCAGCAGUAGCCGCCUCGGCGGCACAGCUGCUGGCGGCCAUGGCGUUGUUGGGAGGCGCAUACUUAGAGAUGUCAAACUUGCGUUUGGGCGUU .((((((...((.((((((((((.(((((....)))))...)))))).)))).))(((((((.....(((.......)))...)))))))......))))))...... ( -42.90) >DroWil_CAF1 2245 108 + 1 GCCAAAGAUUUGACAGCUGCGGUUGCCGCUUCAGCGGCACAACUGCUGGCCGCCUUUCCAUUAUUGGGUGGUGCAUACUUGGAGAUGUCAAAUUUACGCUUCGGCGGC (((..((((((((((((((((((((((((....))))))..))))).)))((((..((((....)))).))))............))))))))))..((....))))) ( -47.20) >DroMoj_CAF1 19190 108 + 1 GCAAGCGAUUUGACCGCUGCAGUAGCUGCCUCUGCGGCACAGCUGCUUGCGGCCAUGCCGUUGUUAGGAGGGGCAUACUUAGAGAGAUCAAACUUGCGUUUGGGUGUU .((((((.(((((.(.(((.(((((((.(((((((((((..((((....))))..)))))).....)))))))).)))))))...).)))))....))))))...... ( -40.10) >DroAna_CAF1 12168 108 + 1 GCCAGGGAUUUGACCGCCGCGGUGGCAGCUUCCGCCGCACAACUGCUGGCCGCCUUCCCAUUGUUCGGCGGCGCAUACUUGGAGAUGUCGAACUUCCGCUUUGGUGUC ((((((((((((((..(((((((((......))))))).....(((..((((((............))))))))).....))....)))))).))))....))))... ( -40.90) >DroPer_CAF1 2154 108 + 1 GCCAGGGAUUUCACAGCCGCUGUCGCCGCUUCGGCGGCACAACUGCUGGCCGCCUUGCCACUGUUCGGUGGCGCAUACUUGGAAAUGUCGAACUUCCGCUUGGGCGUC (((.((((.(((.((((.(.(((.(((((....)))))))).).))))((((((..((....))..))))))((((........)))).))).)))).....)))... ( -46.10) >consensus GCCAGCGAUUUGACCGCAGCAGUAGCCGCUUCGGCGGCACAACUGCUGGCCGCCUUGCCAUUGUUCGGUGGCGCAUACUUGGAGAUGUCAAACUUCCGCUUGGGCGUC (((.(((((((((((.(((((((.(((((....)))))...)))))..((((((............))))))........))...))))))).)))).....)))... (-27.86 = -30.82 + 2.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:35 2006