| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,767,562 – 1,767,664 |
| Length | 102 |
| Max. P | 0.536550 |

| Location | 1,767,562 – 1,767,664 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.76 |
| Mean single sequence MFE | -33.59 |
| Consensus MFE | -19.94 |
| Energy contribution | -21.00 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
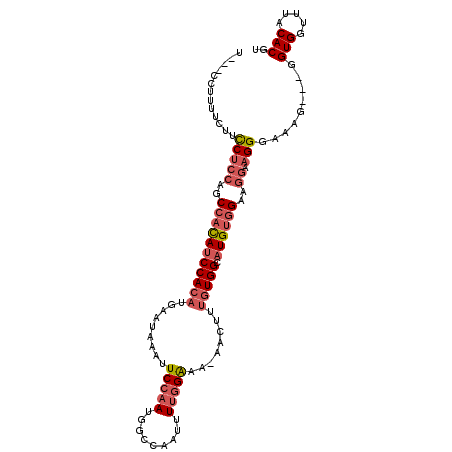
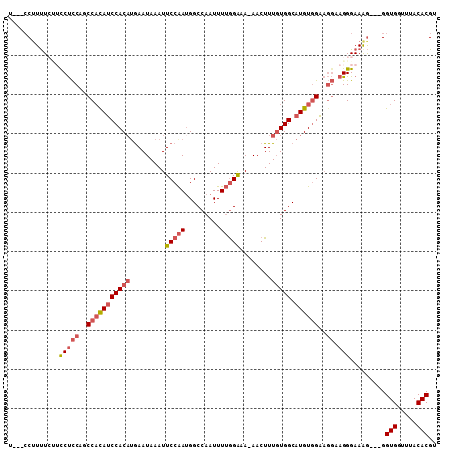
>2L_DroMel_CAF1 1767562 102 + 22407834 U---CCUUUUCUUCCUCCAGCCACAUCCACAUGAAUAAAUUCCAAUGGCCAAUUUUAGGAA-AACUUUGUGGCAUGUGGAAGGAAGGGAAGG---GGUGGUUUACACGU (---((((..((((((....(((((((((((........((((..............))))-.....))))).)))))).))))))..))))---)(((.....))).. ( -37.66) >DroSec_CAF1 24592 100 + 1 U---CCUUUU---CCUCCAGCCACAUCCACAUGAAUAAAUUCCGAAGGCCAAUUUGGGGAAAAACUUUGUGGCAUGUGGAAGGCAGGGAAAG---GGUGGUUUACACGU .---((((((---(((((..(((((((((((........(((((((......)))))))........))))).))))))..)).))))))))---)(((.....))).. ( -41.09) >DroSim_CAF1 26898 99 + 1 U---CCUUUU---UCUCCAGCCAUAUCCACAUGAAUAAAUUCCAAUGGCCAAUUUUGGGAA-AACUUUGUGGCAUGUGGAAGGCAGGGAAAG---GGUGGUUUACACGU .---(((((.---.((((..(((((((((((........((((((.........)))))).-.....))))).))))))..)).))..))))---)(((.....))).. ( -34.94) >DroEre_CAF1 24395 105 + 1 CCUUACUUUUCUUCCUCCAGCCACAUCCACAUGAAUAAAUUCCAAUGGCCAAUUUUGGACA-AACUUUGUGGCAUGUGGAAGGAAGGGAAAG---GGUGGUUUACACGU ((((...(..((((((....(((((((((((.........(((((.........)))))..-.....))))).)))))).))))))..))))---)(((.....))).. ( -35.39) >DroYak_CAF1 33566 102 + 1 U---CCAUUUCUUUCUCCAGCCACAUCCACAUGAAUAAAUUCCAAUGGCCAAUUUUGGACA-AACUUUGUGGCAUGUCGAAGGAAGGGAAAG---GGUGCUUUACACGU (---((.((((((((..((((((((.(((...(((....)))...)))(((....)))...-.....)))))).))..)))))))))))...---.(((.....))).. ( -27.30) >DroAna_CAF1 23805 100 + 1 ----CCUUCUCCUCCCUCUGCCCCAGCCACUCAAAUAAAUUCCAAUGGCCAAUUUUGGACA-AACUUUCUGGCAUGUGGC----AGGGAAAAGUUGGUGGCUUACACGU ----...........(((((((.((((((...........(((((.........)))))..-.......)))).)).)))----))))........(((.....))).. ( -25.15) >consensus U___CCUUUUCUUCCUCCAGCCACAUCCACAUGAAUAAAUUCCAAUGGCCAAUUUUGGAAA_AACUUUGUGGCAUGUGGAAGGAAGGGAAAG___GGUGGUUUACACGU .............(((((..(((((((((((.........(((((.........)))))........))))).))))))..)).))).........(((.....))).. (-19.94 = -21.00 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:02 2006