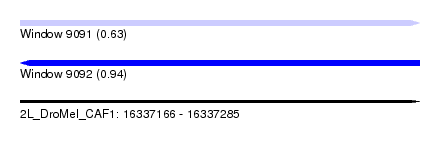
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,337,166 – 16,337,285 |
| Length | 119 |
| Max. P | 0.942197 |

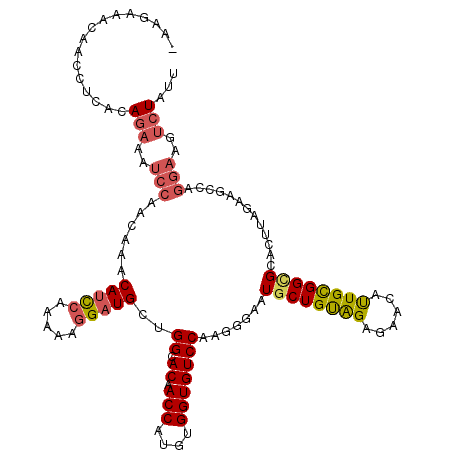
| Location | 16,337,166 – 16,337,285 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -16.80 |
| Energy contribution | -17.92 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16337166 119 + 22407834 AAUAGACUUCCUGGCUUCUAAGUGCGCCGCAAUGAUCUCUACAGCAUUCCCUUGGACACCACAUGGUUGUGCCAGCAUCCUUUUUGGAUGUUUGCUGGAUUUCUGAGAGCUUGUUUCUU- ..((((..(((.(((..........)))((((...........((((..((.((.......)).))..))))..((((((.....)))))))))).)))..))))..............- ( -30.30) >DroSec_CAF1 11074 119 + 1 AAUAGACAUCCUGGCUUUUAAGUGCGCCGCAAUGAUCUCUACAGCAUUCUCUUGGACACCACAUGGUUGUGCCAGCAUCCUUUUUGGAUGUUUGUUGGAUUUCUGUGAGGUUAUUCCUA- ............(((..........)))(.(((((((((.((((...(((..((((((((....)).))).)))((((((.....)))))).....)))...))))))))))))).)..- ( -32.20) >DroSim_CAF1 11701 119 + 1 AAUAGACAUCCUGGCUUUUAAGUGCGCCGCAAUGAUCUCUACAGCAUUCUCUUGGACACCACAUGGUUGUGCCAGCAUCCUUUUUGGAUGUUUGUUGGAUUUCUGUGAGGUUAUUCCUA- ............(((..........)))(.(((((((((.((((...(((..((((((((....)).))).)))((((((.....)))))).....)))...))))))))))))).)..- ( -32.20) >DroEre_CAF1 11572 119 + 1 AACAGACUUCCUGCCUUCUAAGCGCGCCGCAAUGUUCUCUGCAGCAUUCCCUUGGACACCACAUGGUUGUGCCGGCAUCCUUUUUGGAUGUUUGCAGGACUUCUGUGAGUUUGUUUCUU- (((((((((((.((((....)).))((.(((........))).))........)))...((((.((((.(((..((((((.....))))))..))).))))..))))))))))))....- ( -39.30) >DroYak_CAF1 12650 116 + 1 AAUAGACUUCCUGCUCCC---GCGCACCACAAUGUUCUCCACAGCAUUCCCUCGGACACCACAUGGUGGUACCGGCAUCCUUUUUGGAUGUAUUUAGGAUUUCUGUGAGUUUAUUUCUU- ((((((((...(((....---..))).............(((((...(((..(((.((((....))))...)))((((((.....)))))).....)))...)))))))))))))....- ( -33.80) >DroAna_CAF1 8844 105 + 1 AACAU-UAUCCAGACUUCUGACCACUCCGCAGCAUUGUCUGCAGCAUUCCAUUGGACACCACAUGGUGGUGCCCUCAUCCCUA---ACUG-----------UCUGACAAGCGGUUCCUGA .....-....(((....)))......((((....(((((.((((.........((.((((((...)))))).)).........---.)))-----------)..)))))))))....... ( -26.11) >consensus AAUAGACAUCCUGGCUUCUAAGCGCGCCGCAAUGAUCUCUACAGCAUUCCCUUGGACACCACAUGGUUGUGCCAGCAUCCUUUUUGGAUGUUUGUUGGAUUUCUGUGAGGUUAUUCCUU_ ..............................((((..(((.((((...(((..((((((((....))).)).)))((((((.....)))))).....)))...)))))))..))))..... (-16.80 = -17.92 + 1.11)

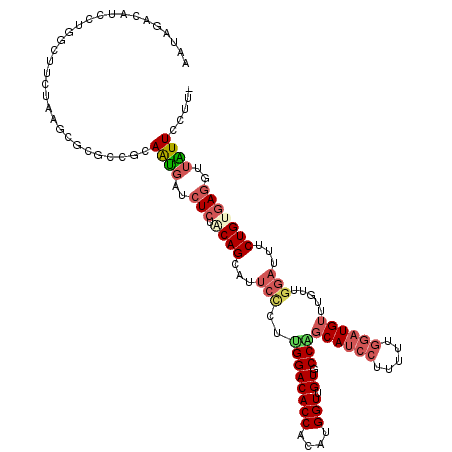

| Location | 16,337,166 – 16,337,285 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -19.25 |
| Energy contribution | -19.70 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16337166 119 - 22407834 -AAGAAACAAGCUCUCAGAAAUCCAGCAAACAUCCAAAAAGGAUGCUGGCACAACCAUGUGGUGUCCAAGGGAAUGCUGUAGAGAUCAUUGCGGCGCACUUAGAAGCCAGGAAGUCUAUU -...............(((..(((.((...(((((.....)))))...)).........((((.((.(((....((((((((......))))))))..))).)).)))))))..)))... ( -30.90) >DroSec_CAF1 11074 119 - 1 -UAGGAAUAACCUCACAGAAAUCCAACAAACAUCCAAAAAGGAUGCUGGCACAACCAUGUGGUGUCCAAGAGAAUGCUGUAGAGAUCAUUGCGGCGCACUUAAAAGCCAGGAUGUCUAUU -.(((.....)))...(((.((((......(((((.....))))).((((...(((....)))......(((..((((((((......))))))))..)))....)))))))).)))... ( -31.30) >DroSim_CAF1 11701 119 - 1 -UAGGAAUAACCUCACAGAAAUCCAACAAACAUCCAAAAAGGAUGCUGGCACAACCAUGUGGUGUCCAAGAGAAUGCUGUAGAGAUCAUUGCGGCGCACUUAAAAGCCAGGAUGUCUAUU -.(((.....)))...(((.((((......(((((.....))))).((((...(((....)))......(((..((((((((......))))))))..)))....)))))))).)))... ( -31.30) >DroEre_CAF1 11572 119 - 1 -AAGAAACAAACUCACAGAAGUCCUGCAAACAUCCAAAAAGGAUGCCGGCACAACCAUGUGGUGUCCAAGGGAAUGCUGCAGAGAACAUUGCGGCGCGCUUAGAAGGCAGGAAGUCUGUU -.............(((((..((((((...(((((.....)))))..((.(((.((....)))))))..(((..((((((((......))))))))..))).....))))))..))))). ( -41.60) >DroYak_CAF1 12650 116 - 1 -AAGAAAUAAACUCACAGAAAUCCUAAAUACAUCCAAAAAGGAUGCCGGUACCACCAUGUGGUGUCCGAGGGAAUGCUGUGGAGAACAUUGUGGUGCGC---GGGAGCAGGAAGUCUAUU -...............(((..((((.....(((((.....)))))((((((((((.((((....((((.((.....)).))))..)))).))))))).)---))....))))..)))... ( -37.20) >DroAna_CAF1 8844 105 - 1 UCAGGAACCGCUUGUCAGA-----------CAGU---UAGGGAUGAGGGCACCACCAUGUGGUGUCCAAUGGAAUGCUGCAGACAAUGCUGCGGAGUGGUCAGAAGUCUGGAUA-AUGUU (((((.((((((..(((..-----------(...---...)..)))(((((((((...))))))))).........((((((......))))))))))))......)))))...-..... ( -34.80) >consensus _AAGAAACAACCUCACAGAAAUCCAACAAACAUCCAAAAAGGAUGCUGGCACAACCAUGUGGUGUCCAAGGGAAUGCUGUAGAGAACAUUGCGGCGCACUUAGAAGCCAGGAAGUCUAUU ................(((..(((......(((((.....)))))..((.((.(((....))))))).......((((((((......)))))))).............)))..)))... (-19.25 = -19.70 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:30 2006