
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,336,912 – 16,337,049 |
| Length | 137 |
| Max. P | 0.852302 |

| Location | 16,336,912 – 16,337,011 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.90 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -18.69 |
| Energy contribution | -17.07 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16336912 99 + 22407834 ---------------------CUGCUCCUCUUCCUCCAUGUAGGAAUAGGUGAGGUUUCCCGCUUCUUGACGAUAAUCGCAACUAGAUCUAUAACUUGAUGUUAGAGGGGAAUCCAGCAA ---------------------.(((((((.(((((......))))).)))...((.(((((.((..(((.((.....)))))..))..((.((((.....)))).))))))).)))))). ( -28.10) >DroSec_CAF1 10823 99 + 1 ---------------------CUGCUCCUCUUCCUCCAUGUAGGAAUAUGUGAGGUUUCCCCCUCCUUGACGAUAAUCGCAGCUAGAUCUAUAACUGGAUGUUCGAGGGGAAUCUAGCAA ---------------------.((((......(((((((((....))))).))))....((((((.(((.((.....)))))((((........))))......)))))).....)))). ( -27.30) >DroSim_CAF1 11450 99 + 1 ---------------------CUGCUCCUCUUCCUCCAUGUAGGAAUAUGUGAGGUUUCCCCCUCCUUGACGAUAAUCGCAGCUAGAUCUAUAACUGGAUGUUCGAGGGGAAUCUAGCAA ---------------------.((((......(((((((((....))))).))))....((((((.(((.((.....)))))((((........))))......)))))).....)))). ( -27.30) >DroEre_CAF1 11330 99 + 1 ---------------------CUGCUCCUCUUCCUCCAUAUAGGAAUAGCUAAGGUUUCCGCCUCCUUGGCGAUAAUCGCGACUAGAUCUGUAACUGGAUGUUUGAGGGGAACCCAGCAG ---------------------(((((((((..(.((((((((((..(((....((((..((((.....))))..))))....)))..))))))..)))).)...))))((...))))))) ( -31.80) >DroYak_CAF1 12396 99 + 1 ---------------------CUGCUCCUCUUCCUCCAUAUAGGAAUAGGUAAGGUUUCCGCCUUCUUGGCGAUAAUCGCAGCUAGAUCUAUAACUUGAUGUUCGAGGGGAACCAGGUAG ---------------------((((((((.(((((......))))).)))...((((((((((.....))).....(((.(((...(((........))))))))).))))))).))))) ( -30.60) >DroAna_CAF1 8544 120 + 1 CGGAUGAGCUUCUUCCCGCUCCUCCUCUUCCUCCUCCACAUAGGAGUAGGUCAGAUGCCCACCUCCACUCUGAAAAUCAGGGGAAGUCACAACAUUAGAGCCCCGUGGGGUGUCCGCCAG .(((.((((........)))).)))(((.(((.((((.....)))).)))..))).(..((((.((((..((.....))((((...((.........)).))))))))))))..)..... ( -39.30) >consensus _____________________CUGCUCCUCUUCCUCCAUAUAGGAAUAGGUGAGGUUUCCCCCUCCUUGACGAUAAUCGCAGCUAGAUCUAUAACUGGAUGUUCGAGGGGAAUCCAGCAA ......................(((((((.(((((......))))).)))...(((((((..((..(((.((.....)))))..))(((........))).......))))))).)))). (-18.69 = -17.07 + -1.63)

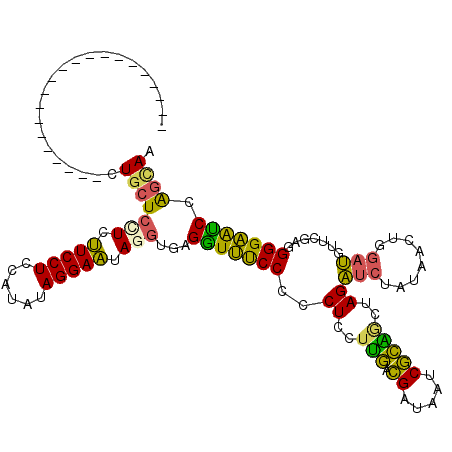
| Location | 16,336,931 – 16,337,049 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.55 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -14.81 |
| Energy contribution | -14.60 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16336931 118 - 22407834 --UCGGGGAGUAUUACCAUCCUACUGGUGAAUCCAUGUACUUGCUGGAUUCCCCUCUAACAUCAAGUUAUAGAUCUAGUUGCGAUUAUCGUCAAGAAGCGGGAAACCUCACCUAUUCCUA --..(((((...((((((......))))))(((((.((....))))))))))))..........((..((((.(((.(..(((.....)))).))).(.((....)).)..))))..)). ( -30.60) >DroSec_CAF1 10842 118 - 1 --ACGGGGAGUAUUACCGUCCUACUGGGGAAUCCAUGUACUUGCUAGAUUCCCCUCGAACAUCCAGUUAUAGAUCUAGCUGCGAUUAUCGUCAAGGAGGGGGAAACCUCACAUAUUCCUA --...((((((((.....((((...((((((((...((....))..)))))))).(((..(((((((((......)))))).)))..)))...))))((((....))))..)))))))). ( -43.20) >DroSim_CAF1 11469 118 - 1 --ACGGGGAGUAUUACCGUCCUACUGGGGAAUCCAUGUACUUGCUAGAUUCCCCUCGAACAUCCAGUUAUAGAUCUAGCUGCGAUUAUCGUCAAGGAGGGGGAAACCUCACAUAUUCCUA --...((((((((.....((((...((((((((...((....))..)))))))).(((..(((((((((......)))))).)))..)))...))))((((....))))..)))))))). ( -43.20) >DroEre_CAF1 11349 118 - 1 --ACGAGGUGUAUUACCAUCCCAGUGGUGAAUCCAUGUAUCUGCUGGGUUCCCCUCAAACAUCCAGUUACAGAUCUAGUCGCGAUUAUCGCCAAGGAGGCGGAAACCUUAGCUAUUCCUA --...(((.((((((((((....)))))))............((((((((((((((.(((.....)))............(((.....)))....)))).)).)))).))))))).))). ( -31.30) >DroYak_CAF1 12415 118 - 1 --ACGAGGAGUAUUACCAUCCUAAGGGAGAAUCCACUUACCUACCUGGUUCCCCUCGAACAUCAAGUUAUAGAUCUAGCUGCGAUUAUCGCCAAGAAGGCGGAAACCUUACCUAUUCCUA --...(((((((.........((((((.....)).)))).......((((.((...((..(((.(((((......)))))..)))..))(((.....))))).)))).....))))))). ( -26.40) >DroAna_CAF1 8584 109 - 1 AUAGGAAGA-----------GUAGUGGACCCUCCAUCUACCUGGCGGACACCCCACGGGGCUCUAAUGUUGUGACUUCCCCUGAUUUUCAGAGUGGAGGUGGGCAUCUGACCUACUCCUA .(((((((.-----------(((((((.....))).))))))((((((..(((..((.(((......))).))((((((((((.....))).).)))))))))..)))).))...))))) ( -37.80) >consensus __ACGAGGAGUAUUACCAUCCUACUGGAGAAUCCAUGUACCUGCUGGAUUCCCCUCGAACAUCCAGUUAUAGAUCUAGCUGCGAUUAUCGUCAAGGAGGGGGAAACCUCACCUAUUCCUA .....(((((((...(((......)))....(((........((((((((.....................)))))))).(((.....))).........))).........))))))). (-14.81 = -14.60 + -0.21)
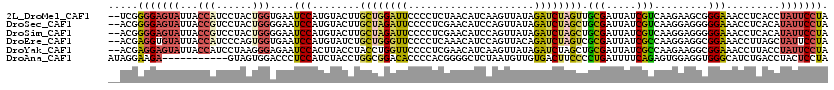

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:28 2006