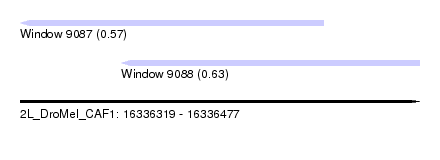
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,336,319 – 16,336,477 |
| Length | 158 |
| Max. P | 0.629542 |

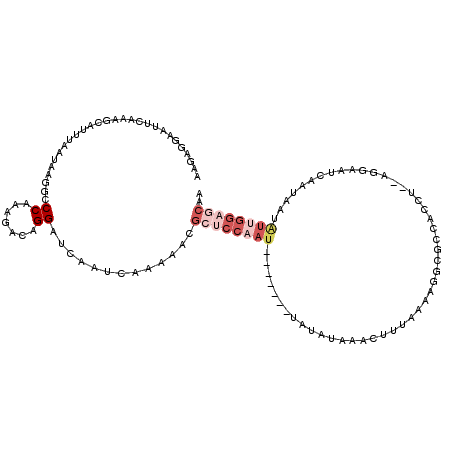
| Location | 16,336,319 – 16,336,439 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.76 |
| Mean single sequence MFE | -17.02 |
| Consensus MFE | -3.41 |
| Energy contribution | -5.29 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.20 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16336319 120 - 22407834 GGUCAAUCAAAAACGCUCCAAUAGUCCAGUAUAUAAACUUUAAAAGGCGCCACCUAAAGGAAUCAAUAAUGUUGGAGCAAAAUGAUCUUAAAGCAAUCUUUUACAGUCCCAUCAAAUUUA ((............(((((((((.((((((......))).....(((.....)))...)))........))))))))).....(((..(((((.....)))))..))))).......... ( -19.90) >DroSec_CAF1 10251 99 - 1 GAUCAAUCAGAAACGCUCCAAU--------AUAUAAACUUUAAAA-------------GGAGUCAAUAAUAUUGGAGCAAAAUGACCUUAAAGCAAUCUUUUACAGUCCCAUCAAAUUUA (((...........((((((((--------((((..(((((....-------------)))))..)).)))))))))).....(((..(((((.....)))))..)))..)))....... ( -19.60) >DroSim_CAF1 10878 99 - 1 GAUCAAUCAGAAACGCUCCAAU--------AUAUAAACUUUAAAA-------------GGAGUCAAUAAUAUUGGAGCAAAAUGACCUUAAAGCAAUCUUUUACAGUCCCAUCAAAUUUA (((...........((((((((--------((((..(((((....-------------)))))..)).)))))))))).....(((..(((((.....)))))..)))..)))....... ( -19.60) >DroEre_CAF1 10749 120 - 1 GAUCAGUCAAAAGCGCUCCGAUAGUCCAAUAUAUAAACAUUAAAAGGCGCCACCUGCAGGAAUAAAUAAUAUGGGUCCAUAAUGAUCUUAAAGCAAUCUCUUACAGUCCUAUCAAACUUA (((.((......(((((............................)))))...(((.((((...........(((((......)))))..........)))).)))..)))))....... ( -13.39) >DroYak_CAF1 11855 101 - 1 GAUCUAUC-------------------GAUCUAUAAACAUAAAAAGGCGCCACCUAGAGGAAUAAAUAAUAUAGAAACAUAAGGACCUUAAAGCAAUCUUUUACAGUCCUAUCAAAUUUA (((.....-------------------..((((((..........(....).((....)).........))))))......(((((..(((((.....)))))..))))))))....... ( -12.60) >consensus GAUCAAUCAAAAACGCUCCAAU_______UAUAUAAACUUUAAAAGGCGCCACCU__AGGAAUCAAUAAUAUUGGAGCAAAAUGACCUUAAAGCAAUCUUUUACAGUCCCAUCAAAUUUA (((...........((((((((................................................)))))))).....(((..(((((.....)))))..)))..)))....... ( -3.41 = -5.29 + 1.88)

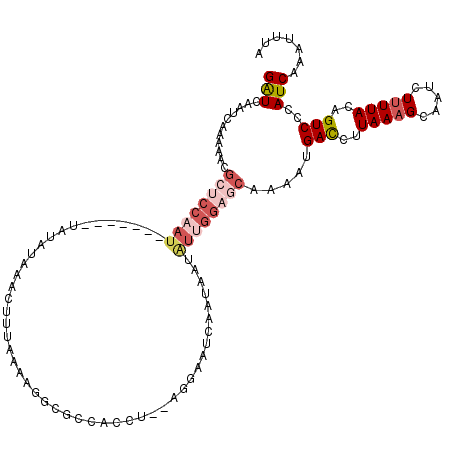
| Location | 16,336,359 – 16,336,477 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.71 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -1.85 |
| Energy contribution | -3.93 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.10 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16336359 118 - 22407834 AAAAGGAAUUCUAAGCAUUUAAUAAGGUCCAAAGACAGGGUCAAUCAAAAACGCUCCAAUAGUCCAGUAUAUAAACUUUAAAAGGCGCCACCUAAAGGAAUCAAUAAUGUUGGAGCAA .........................(..((.......))..)..........(((((((((.((((((......))).....(((.....)))...)))........))))))))).. ( -22.60) >DroSec_CAF1 10291 97 - 1 AAGAGGAAUUCAAAGCAUUUAAUAAGGCCCAAAGACAGGAUCAAUCAGAAACGCUCCAAU--------AUAUAAACUUUAAAA-------------GGAGUCAAUAAUAUUGGAGCAA .........................(..((.......))..)..........((((((((--------((((..(((((....-------------)))))..)).)))))))))).. ( -19.40) >DroSim_CAF1 10918 97 - 1 AAGAGGAAUUCAAAGCAUUUAAUAAGGCCCAAAGACAGGAUCAAUCAGAAACGCUCCAAU--------AUAUAAACUUUAAAA-------------GGAGUCAAUAAUAUUGGAGCAA .........................(..((.......))..)..........((((((((--------((((..(((((....-------------)))))..)).)))))))))).. ( -19.40) >DroEre_CAF1 10789 118 - 1 AAGGGAAGUUCAAAGCAUCGAAUAAAGCCCAAGGACAGGAUCAGUCAAAAGCGCUCCGAUAGUCCAAUAUAUAAACAUUAAAAGGCGCCACCUGCAGGAAUAAAUAAUAUGGGUCCAU ..((...((((........))))...(((((.((((.(((...((.....))..)))....)))).................(((.....)))................))))))).. ( -20.10) >DroYak_CAF1 11895 99 - 1 AAGGAGAAUUCAAAGCACCUAAUAAAGCCCAAAGACAGGAUCUAUC-------------------GAUCUAUAAACAUAAAAAGGCGCCACCUAGAGGAAUAAAUAAUAUAGAAACAU ........(((...((.(((......(........).(((((....-------------------)))))............))).))..((....)).............))).... ( -10.00) >consensus AAGAGGAAUUCAAAGCAUUUAAUAAGGCCCAAAGACAGGAUCAAUCAAAAACGCUCCAAU_______UAUAUAAACUUUAAAAGGCGCCACCU__AGGAAUCAAUAAUAUUGGAGCAA ............................((.......)).............((((((((................................................)))))))).. ( -1.85 = -3.93 + 2.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:26 2006