
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,333,433 – 16,333,547 |
| Length | 114 |
| Max. P | 0.519592 |

| Location | 16,333,433 – 16,333,547 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.94 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -6.20 |
| Energy contribution | -6.23 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.26 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
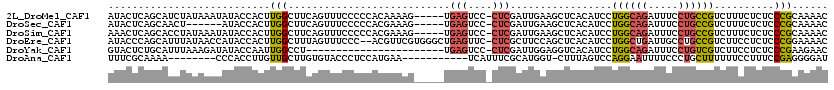
>2L_DroMel_CAF1 16333433 114 + 22407834 AUACUCAGCAUCUAUAAAUAUACCACUUGGCUUCAGUUUCCCCCACAAAAG-----UGAGUCC-CUCGAUUGAAGCUCACAUCCUGGCAGAUUUCCUGCCGUCUUUCUCUCCCGCAAAAC .......((...................((((((((((....((((....)-----)).)...-...))))))))))........(((((.....))))).............))..... ( -22.80) >DroSec_CAF1 7364 108 + 1 AUACUCAGCAACU------AUACCACUUGGCUUCAGUUUCCCCCACGAAAG-----UGAGUCC-CUCGAUUGAAGCUCACAUCCUGGCAGAUUUCCUGCCGUCUUUCUCUCCCGCAAAAC .......((....------.........((((((((((....((((....)-----)).)...-...))))))))))........(((((.....))))).............))..... ( -26.50) >DroSim_CAF1 7975 114 + 1 AAACUCAGCACCUAUAAAUAUACCACUUGGCUUCAGUUUCCCCCACGAAAG-----UGAGUCC-CUCGAUUGAAGCUCACAUCCUGGCAGAUUUCCUGCCGUCUUUCUCUCCCGCAAAAC .......((...................((((((((((....((((....)-----)).)...-...))))))))))........(((((.....))))).............))..... ( -26.50) >DroEre_CAF1 7590 117 + 1 AUACCCAGCAUUUAUAACCAUACCACUUGGCUUUAGUUUCCC--ACGUUCGUGGGCUGAGUUC-CUCGCUUCCAGCUCACAUCCUGGCUGAUUGCCUGCCGUCUUCCUCUCCCGGAAAAC ............................(((..(((((..((--(.(...(((((((((((..-...)))..))))))))..).)))..)))))...)))((.((((......)))).)) ( -26.80) >DroYak_CAF1 8278 96 + 1 GUACUCUGCAUUUAAAGAUAUACCAAUUGGCCU-----------------------UGAGUCC-CUCGAUUGGAGGUCACAUCCUGGCAGAUUUCCUGUCGUCUUCCUCUCCCGAAGAAC (((.(((........)))..)))....((((((-----------------------(.((((.-...)))).)))))))......(((((.....))))).(((((.......))))).. ( -22.80) >DroAna_CAF1 5087 100 + 1 UUUCGCAAAA--------CCCACCUUGUUGCUUGUGUACCCUCCAUGAA-----------UCAUUUCGCAUGGU-CUUUAGUCCAGGAAUUUUCCCUGCUUUUUUCCUUUCCGAGGGGAU ...(((((((--------(.......)))..)))))..(((((...(((-----------....)))((((((.-(....).)))(((....))).))).............)))))... ( -18.50) >consensus AUACUCAGCAUCUAUAAAUAUACCACUUGGCUUCAGUUUCCCCCACGAAAG_____UGAGUCC_CUCGAUUGAAGCUCACAUCCUGGCAGAUUUCCUGCCGUCUUCCUCUCCCGCAAAAC ...........................(((...........................(((....))).................((((((.....))))))..........)))...... ( -6.20 = -6.23 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:14 2006