| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,765,508 – 1,765,609 |
| Length | 101 |
| Max. P | 0.775201 |

| Location | 1,765,508 – 1,765,609 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.52 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
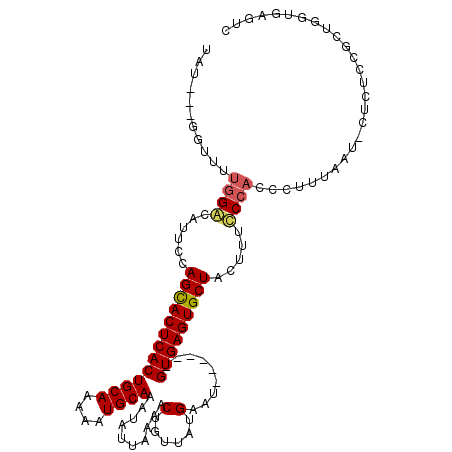
>2L_DroMel_CAF1 1765508 101 + 22407834 UAU---CGUUUUGGGCAUUCCAGUACUCACUGCAAAAAUGCAAAUAUUAAAACU---AUGAAU------GUGAGUGCUCUUUUCCCACCCUUUAAA-CUUUCAGCUAGUGAGUC ...---.....((((......(((((((((((((....))))....(((.....---.)))..------))))))))).....))))........(-(((.(.....).)))). ( -19.10) >DroSec_CAF1 22564 105 + 1 UAU---GGUUUUGGGCAUUCCAGCACUCACUGCAAAAAUGCAAAUAUUAAAACUGUUAUGAGU------GUGAGUGCUCCUUUCCCACCCUUUAAUUCUCUCUGCUGGCGAGUC ...---((...((((......(((((((((((((....)))).........(((......)))------))))))))).....)))).)).......(((((....)).))).. ( -25.40) >DroEre_CAF1 22330 86 + 1 U-----GGGUUUGGACAUUCCAGCACUCACUGCAAAAAUGCAAAUAUUAAAACUGUUAUGAAU------GUGAGUGCUACUUUCC-----------------CGCUGGUGAGUC .-----(((...((.....))(((((((((((((....))))((((.......))))......------))))))))).....))-----------------)........... ( -22.50) >DroYak_CAF1 31454 114 + 1 UCCCGUAUAUUUGGACAUUCCAGCACUCACUGCACAAAUGCAAAUAUUAAAACUGUUAUGAAUGCGAGUGUGAGUGCUACUUUUCCACACUUUAAUCCUCACCGCUGGUGAGUC ...........((((......(((((((((((((....)))).........(((((.......)).)))))))))))).....))))..........((((((...)))))).. ( -29.00) >consensus UAU___GGUUUUGGACAUUCCAGCACUCACUGCAAAAAUGCAAAUAUUAAAACUGUUAUGAAU______GUGAGUGCUACUUUCCCACCCUUUAAU_CUCUCCGCUGGUGAGUC ...........((((......(((((((((((((....))))..........(......).........))))))))).....))))........................... (-17.36 = -17.68 + 0.31)

| Location | 1,765,508 – 1,765,609 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.52 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -17.89 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
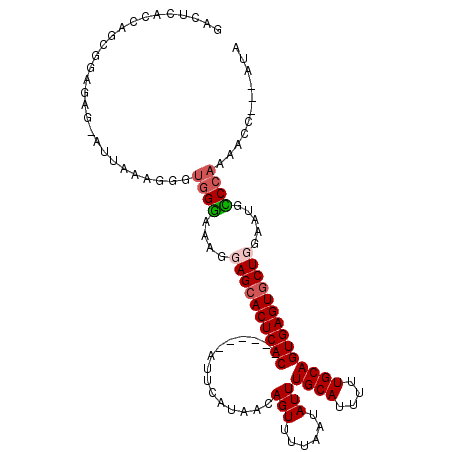
>2L_DroMel_CAF1 1765508 101 - 22407834 GACUCACUAGCUGAAAG-UUUAAAGGGUGGGAAAAGAGCACUCAC------AUUCAU---AGUUUUAAUAUUUGCAUUUUUGCAGUGAGUACUGGAAUGCCCAAAACG---AUA .(((((((.((.(((((-(.(((((((((.........)))))((------......---.)).......)))).)))))))))))))))..(((.....))).....---... ( -19.40) >DroSec_CAF1 22564 105 - 1 GACUCGCCAGCAGAGAGAAUUAAAGGGUGGGAAAGGAGCACUCAC------ACUCAUAACAGUUUUAAUAUUUGCAUUUUUGCAGUGAGUGCUGGAAUGCCCAAAACC---AUA ..(((.......))).........((.((((.....(((((((((------(((......))).........((((....)))))))))))))......))))...))---... ( -30.80) >DroEre_CAF1 22330 86 - 1 GACUCACCAGCG-----------------GGAAAGUAGCACUCAC------AUUCAUAACAGUUUUAAUAUUUGCAUUUUUGCAGUGAGUGCUGGAAUGUCCAAACCC-----A ...........(-----------------((.....(((((((((------.....(((.....))).....((((....)))))))))))))((.....))...)))-----. ( -24.40) >DroYak_CAF1 31454 114 - 1 GACUCACCAGCGGUGAGGAUUAAAGUGUGGAAAAGUAGCACUCACACUCGCAUUCAUAACAGUUUUAAUAUUUGCAUUUGUGCAGUGAGUGCUGGAAUGUCCAAAUAUACGGGA ..((((((...))))))..........((((....(((((((((((((............))).........((((....)))))))))))))).....))))........... ( -30.90) >consensus GACUCACCAGCGGAGAG_AUUAAAGGGUGGGAAAGGAGCACUCAC______AUUCAUAACAGUUUUAAUAUUUGCAUUUUUGCAGUGAGUGCUGGAAUGCCCAAAACC___AUA ...........................((((....((((((((((...............(((......)))((((....)))))))))))))).....))))........... (-17.89 = -18.70 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:01 2006