| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,309,277 – 16,309,383 |
| Length | 106 |
| Max. P | 0.507011 |

| Location | 16,309,277 – 16,309,383 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.35 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -21.93 |
| Energy contribution | -20.85 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
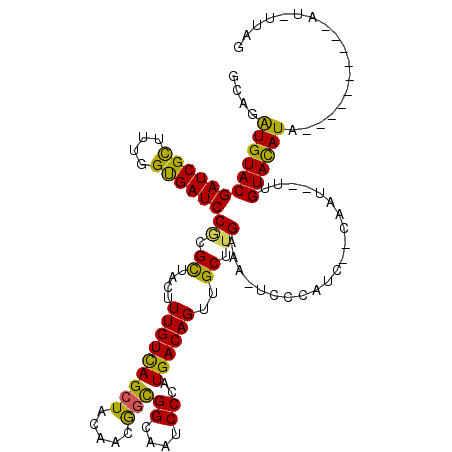
>2L_DroMel_CAF1 16309277 106 - 22407834 GCAGAUGUACGAUCGCUUUGGUGAUCCUCGCUAUUUUGUCAGCUACAACGGUGGCAAUCCCAUGACAGUUGCCUGAAUG-CCCAUC--CAAU--CUGUACAUACG--------AU-UUAG ....((((((((((((....))))))...((....(((((((((((....))))).......))))))..)).......-......--....--..))))))...--------..-.... ( -24.71) >DroVir_CAF1 9761 98 - 1 GCAGGUGUACGAUCGCUUCGGUGAUCCGCGCUACUUUGUCAGCUAUAAUGGCGGCAAUCCCAUGACAGUUGCUUGAAA-UUGCAUCAACAA---UUGUACAU------------------ ...(((((..((((((....)))))).)))))...(((((.(((.....))))))))....((((((((((.((((..-.....)))))))---)))).)))------------------ ( -30.40) >DroGri_CAF1 10103 105 - 1 GCAGAUGUACGAUCGCUUUGGUGAUCCGCGUUACUUUGUCAGCUACAAUGGCGGCAAUCCGAUGACAGUUGCUUGAGA-UCGCAUCAUCAA---UUGUACAUA----------AU-AUAU ....(((((((((.......((((((((.((....(((((((((.....)))((....))..))))))..)).)).))-)))).......)---)))))))).----------..-.... ( -27.84) >DroWil_CAF1 13313 107 - 1 ACAGAUGUACGAUCGCUUUGGCGAUCCACGUUACUUUGUUAGCUAUGGCAAUGGCAAUCCCAUGACAGUUGCUUGAAACUUGCAUC--CAUUAAUUGUAUAUA----------AU-AUAC ...((((((.((((((....))))))((.((....((((((((....))..(((.....)))))))))..)).)).....))))))--...............----------..-.... ( -22.20) >DroAna_CAF1 7365 108 - 1 GCAGAUGUACGAUCGCUUCGGUGAUCCGCGUUAUUUUGUCAGCUACAACGGCGGCAAUCCAAUGACAGUUGCUUGAAUAACCCAAC--CACU--UUGUACAUACA--------ACCUUAG ....((((((((((((....)))))).(.(((((((((((((((.....)))((....))..))))).......))))))).)...--....--..))))))...--------....... ( -27.41) >DroPer_CAF1 7048 114 - 1 GCAGAUGUACGAUCGUUUUGGUGAUCCGCGCUACUUUGUCAGCUACAACGGCGGCAAUCCCAUGACAGUUGCCUGAGUG-CCCCAA--CCAC--UUGUACAUACCACCCCAUAAC-UUAG ....((((((((......((((.....(((((.........(((.....)))((((((.........))))))..))))-)....)--))).--)))))))).............-.... ( -25.10) >consensus GCAGAUGUACGAUCGCUUUGGUGAUCCGCGCUACUUUGUCAGCUACAACGGCGGCAAUCCCAUGACAGUUGCUUGAAA_UCCCAUC__CAAU__UUGUACAUA__________AU_UUAG ....((((((((((((....))))))((.((....(((((((((.....)))((....))..))))))..)).)).....................)))))).................. (-21.93 = -20.85 + -1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:09 2006