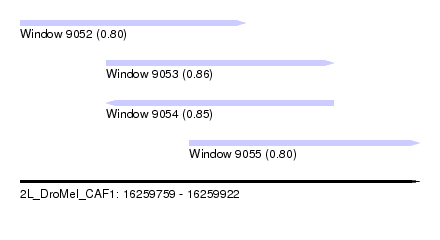
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,259,759 – 16,259,922 |
| Length | 163 |
| Max. P | 0.862893 |

| Location | 16,259,759 – 16,259,851 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -23.01 |
| Energy contribution | -23.07 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16259759 92 + 22407834 GAAGAAGAGUUUC-UUCAAUU---CAAAUC-CGGGUCUCAACA--GUGGCGUCCGAGCAGGGACACUUGGUGUCCUAGCUGGCAGCUAUGAGACGUGUU ((((((....)))-)))....---......-((.((((((...--.((((..((.(((.(((((((...))))))).)))))..)))))))))).)).. ( -34.40) >DroSec_CAF1 1176 93 + 1 GAACAAGAGCUUC-UUCAAUU---CGAAUCGCGGGUCUCAACA--GUGGCGCCCGAGCAGGGACACUUGGUGUCCUAGCUGGCAGCUAUGAGACGUCUU ....((((((...-(((....---.)))..))..((((((...--.(((((((..(((.(((((((...))))))).)))))).)))))))))).)))) ( -32.40) >DroSim_CAF1 1178 93 + 1 GAACAAGAGCUUC-UUCAAUU---CGAAUCGCGGGUCUCAACA--GUGGCGCCCCAGCAGGGACACUUGGUGUCCUAGCUGGCAGCUAUGAGACGUCUU ....((((((...-(((....---.)))..))..((((((...--.((((...(((((.(((((((...))))))).)))))..)))))))))).)))) ( -34.60) >DroEre_CAF1 1166 91 + 1 GAAGAAGAGUUUC-UUCAAUU---CGAAUC--GGGUCUCUACA--GUGGCGCCCGAGCAGGGACACUUGGUGUCCUAGCUGGCAGCUAUAAGACGUUUU ((((((....)))-)))....---......--..((((.....--((((((((..(((.(((((((...))))))).)))))).))))).))))..... ( -30.50) >DroYak_CAF1 1212 91 + 1 AAAGAAGAGCUUC-UUCAUUU---CGAAUC--GGGUCUCUACA--GUGGCGCCCGAGCAGGAACACUUGGUGUCCUAGCUGGUAGCCAUGACACUUGUU ...((((.....)-)))....---.....(--((((.((....--(((((..((.(((((((.(((...))))))).)))))..))))))).))))).. ( -27.50) >DroAna_CAF1 2139 88 + 1 GAAU---GGCUUCCCACCAUCCUACGAUG--CUGGUCA-AAUGGUCUGGCGCCUGAGCAG-GACACUUGGUGUCCUAGAUGGCUGCUGGCUGCCG---- ....---(((...(((.((((....))))--.)))...-...((.(..(((((....(((-(((((...))))))).)..))).))..)))))).---- ( -31.80) >consensus GAAGAAGAGCUUC_UUCAAUU___CGAAUC_CGGGUCUCAACA__GUGGCGCCCGAGCAGGGACACUUGGUGUCCUAGCUGGCAGCUAUGAGACGUCUU ..................................(((((......((((((((..(((.(((((((...))))))).)))))).))))))))))..... (-23.01 = -23.07 + 0.06)

| Location | 16,259,794 – 16,259,887 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -20.64 |
| Energy contribution | -20.03 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.862893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16259794 93 + 22407834 ACA--GUGGCGUCCGAGCAGGGACACUUGGUGUCCUAGCUGGCAGCUAUGAGACGUGUUGAACAAA--GUCAAGAAGAAACUGAAAUUGCACAGCGC ...--(((((..((.(((.(((((((...))))))).)))))..))))).....((((((..(((.--.(((.........)))..)))..)))))) ( -30.10) >DroSec_CAF1 1212 95 + 1 ACA--GUGGCGCCCGAGCAGGGACACUUGGUGUCCUAGCUGGCAGCUAUGAGACGUCUUGAACGAAACGUCAAGAAGAAACUGAAAUUGCACGGCGC ...--...(((((..(((.(((((((...))))))).))).((((...(.((...((((((.((...)))))))).....)).)..))))..))))) ( -31.30) >DroSim_CAF1 1214 95 + 1 ACA--GUGGCGCCCCAGCAGGGACACUUGGUGUCCUAGCUGGCAGCUAUGAGACGUCUUGAACAAAACGUCAAGAAGAAACUGACAUUGCACAGCGC ...--...((((.(((((.(((((((...))))))).)))))..((.(((.(((((.((....)).))))).((......))..))).))...)))) ( -33.30) >DroEre_CAF1 1200 90 + 1 ACA--GUGGCGCCCGAGCAGGGACACUUGGUGUCCUAGCUGGCAGCUAUAAGACGUUUUGAAUAAA--GUUA---AGAAACUGAAAUUGCACAGCGC ...--((((((((..(((.(((((((...))))))).)))))).)))))....((((.(((((..(--(((.---...))))...))).)).)))). ( -28.30) >DroYak_CAF1 1246 90 + 1 ACA--GUGGCGCCCGAGCAGGAACACUUGGUGUCCUAGCUGGUAGCCAUGACACUUGUUGAACAAA--GUUA---AGAAACUGAAAUUGCACAGCGC ...--(((((..((.(((((((.(((...))))))).)))))..))))).......((((..(((.--.(((---......)))..)))..)))).. ( -25.30) >DroAna_CAF1 2173 81 + 1 AUGGUCUGGCGCCUGAGCAG-GACACUUGGUGUCCUAGAUGGCUGCUGGCUGCCG----GAAU----C-------GGCUGUUGGAAUCGCUCAGCGU ........((((.(((((((-(((((...))))))).(((.....(..((.((((----....----)-------))).))..).)))))))))))) ( -36.20) >consensus ACA__GUGGCGCCCGAGCAGGGACACUUGGUGUCCUAGCUGGCAGCUAUGAGACGUCUUGAACAAA__GUCA___AGAAACUGAAAUUGCACAGCGC .....((((((((..(((.(((((((...))))))).)))))).)))))...............................(((........)))... (-20.64 = -20.03 + -0.61)


| Location | 16,259,794 – 16,259,887 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16259794 93 - 22407834 GCGCUGUGCAAUUUCAGUUUCUUCUUGAC--UUUGUUCAACACGUCUCAUAGCUGCCAGCUAGGACACCAAGUGUCCCUGCUCGGACGCCAC--UGU (((.(((((((..((((.......)))).--.))))...))))))......((..(((((..((((((...))))))..))).))..))...--... ( -23.80) >DroSec_CAF1 1212 95 - 1 GCGCCGUGCAAUUUCAGUUUCUUCUUGACGUUUCGUUCAAGACGUCUCAUAGCUGCCAGCUAGGACACCAAGUGUCCCUGCUCGGGCGCCAC--UGU (((((.........(((((.......(((((((......)))))))....)))))(.(((..((((((...))))))..))).))))))...--... ( -32.80) >DroSim_CAF1 1214 95 - 1 GCGCUGUGCAAUGUCAGUUUCUUCUUGACGUUUUGUUCAAGACGUCUCAUAGCUGCCAGCUAGGACACCAAGUGUCCCUGCUGGGGCGCCAC--UGU (((((.........(((((.......((((((((....))))))))....)))))(((((..((((((...))))))..))))))))))...--... ( -38.70) >DroEre_CAF1 1200 90 - 1 GCGCUGUGCAAUUUCAGUUUCU---UAAC--UUUAUUCAAAACGUCUUAUAGCUGCCAGCUAGGACACCAAGUGUCCCUGCUCGGGCGCCAC--UGU ((((..........(((((...---..((--(((.....))).)).....)))))(((((..((((((...))))))..))).))))))...--... ( -23.20) >DroYak_CAF1 1246 90 - 1 GCGCUGUGCAAUUUCAGUUUCU---UAAC--UUUGUUCAACAAGUGUCAUGGCUACCAGCUAGGACACCAAGUGUUCCUGCUCGGGCGCCAC--UGU (((((.((.(((...((((...---.)))--)..)))))...)))))..((((..(((((..((((((...))))))..))).))..)))).--... ( -25.70) >DroAna_CAF1 2173 81 - 1 ACGCUGAGCGAUUCCAACAGCC-------G----AUUC----CGGCAGCCAGCAGCCAUCUAGGACACCAAGUGUC-CUGCUCAGGCGCCAGACCAU ..((((.(((....)....(((-------(----....----)))).)))))).(((...((((((((...)))))-)))....))).......... ( -29.40) >consensus GCGCUGUGCAAUUUCAGUUUCU___UGAC__UUUGUUCAACACGUCUCAUAGCUGCCAGCUAGGACACCAAGUGUCCCUGCUCGGGCGCCAC__UGU ((((((.(((.......................................((((.....))))((((((...)))))).))).))).)))........ (-18.58 = -18.72 + 0.14)


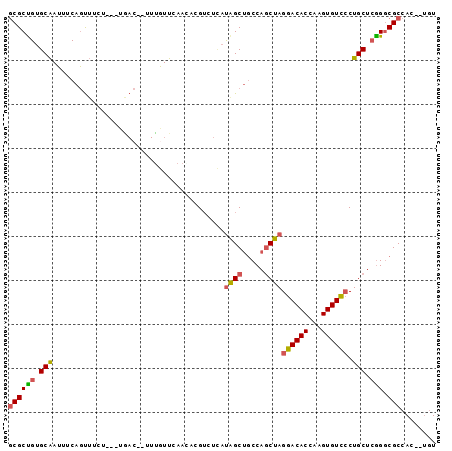
| Location | 16,259,828 – 16,259,922 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -16.65 |
| Energy contribution | -17.18 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16259828 94 + 22407834 AGCUGGCAGCUAUGAGACGUGUUGAACAAA--GUCAAGAAGAAACUGAAAUUGCACAGCGCAGACGCAGCUUGGCGAAG-GCAGACAGCUGCAGCAU--- .((((.(((((....(((.((.....))..--))).........(((...((((..(((((....)).)))..))))..-.)))..)))))))))..--- ( -28.80) >DroSec_CAF1 1246 96 + 1 AGCUGGCAGCUAUGAGACGUCUUGAACGAAACGUCAAGAAGAAACUGAAAUUGCACGGCGCAGACGCAGCCUGGCGAAG-GGAGACAGCUGCAGCAU--- .((((.(((((....(((((.((....)).)))))...............((((..(((((....)).)))..))))..-(....))))))))))..--- ( -32.10) >DroSim_CAF1 1248 96 + 1 AGCUGGCAGCUAUGAGACGUCUUGAACAAAACGUCAAGAAGAAACUGACAUUGCACAGCGCAGACGCAGCUUGGCGAAG-GGAGACAGCUGCAGCAU--- .((((.(((((....(((((.((....)).))))).........((..(.((((..(((((....)).)))..)))).)-..))..)))))))))..--- ( -31.20) >DroEre_CAF1 1234 91 + 1 AGCUGGCAGCUAUAAGACGUUUUGAAUAAA--GUUA---AGAAACUGAAAUUGCACAGCGCAGACGCAGCUUGGCGAAG-GGAGACAGCCGCUGCAU--- .((((((((.....((...((((((.....--.)))---)))..))....)))).))))......(((((..(((....-(....).))))))))..--- ( -27.70) >DroYak_CAF1 1280 91 + 1 AGCUGGUAGCCAUGACACUUGUUGAACAAA--GUUA---AGAAACUGAAAUUGCACAGCGCAGACGCAGCUUGGCGAAG-GGAGACAGCUGCAACAU--- .((((((((....(((....)))......(--(((.---...))))....)))).))))((((.(((......)))...-(....)..)))).....--- ( -20.70) >DroAna_CAF1 2208 85 + 1 AGAUGGCUGCUGGCUGCCG----GAAU----C-------GGCUGUUGGAAUCGCUCAGCGUCGGCGCGGCUUGGCAGAGGAGAGGCAGCUGCGGCAGAUC .(((.(((((.(((((((.----...(----(-------..((((..(....(((..(((....)))))))..))))..))..))))))))))))..))) ( -37.60) >consensus AGCUGGCAGCUAUGAGACGUCUUGAACAAA__GUCA___AGAAACUGAAAUUGCACAGCGCAGACGCAGCUUGGCGAAG_GGAGACAGCUGCAGCAU___ .((((.(((((.......................................((((..(((((....)).)))..))))...(....))))))))))..... (-16.65 = -17.18 + 0.53)

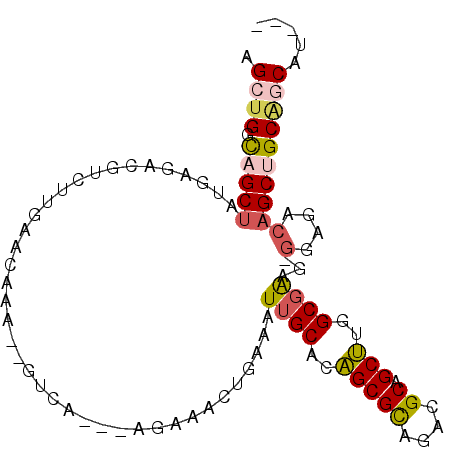
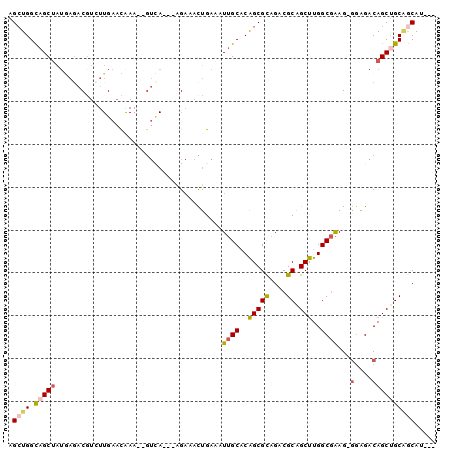
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:54 2006