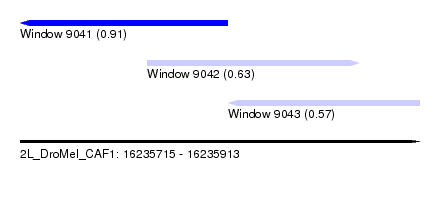
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,235,715 – 16,235,913 |
| Length | 198 |
| Max. P | 0.913770 |

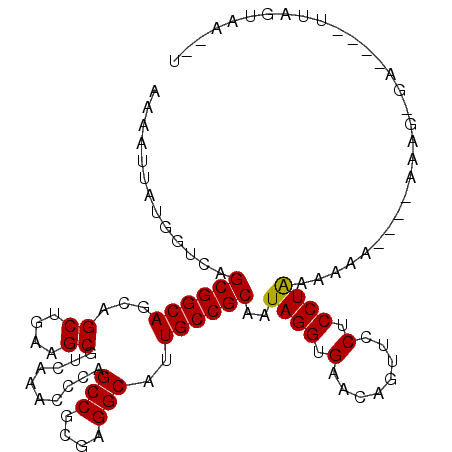
| Location | 16,235,715 – 16,235,818 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -24.79 |
| Energy contribution | -24.65 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16235715 103 - 22407834 AAAAUUAUGGUCAGCGGCAUUAGCUGAAGCGUCAAAACCAGCCGCGAGGCAUUGCCGCAAUAGGUGUACAGUUCCUCCUAAAUAA----GAAG-UAAAUAUUAGUAU--U ....((((..((.((((((...((....))..........(((....)))..))))))..((((.(........).)))).....----)).)-)))..........--. ( -26.30) >DroVir_CAF1 219197 98 - 1 AAAAGUAUGGUCAGCGGCAGCAGCUGAAGCGUCAGGCCCAGCCGUGAGGCGUUGCCGCAAUAGGUGAACAGUUCCUCCUAAAAAA---AACA--------UUUAUGA-AU ..........(((((((((((..((((....)))).....(((....)))))))))))..((((.(........).)))).....---....--------....)))-.. ( -32.50) >DroGri_CAF1 166454 105 - 1 AAAAUAAUGGUCAGCGGCAGCAGCUGAAGCGUCAAACCCAGCCGCGAGGCAUUGCCGCAAUAGGUGAACAGUUCCUCCUACAACA---AAAAGGGUGUC-AUUAUCA-UU ...(((((((.(((((((((..((....))..........(((....))).)))))))..((((.(........).)))).....---.......))))-)))))..-.. ( -29.70) >DroEre_CAF1 156815 102 - 1 AAAAUUAUGGUCAGCGGCAUUAGCUGAAGCGUCAAACCCAGCCGCGAGGCAUUGCCGCAAUAGGUGUACAGUUCCUCCUAAAUAA----AAAU-GAAUCGA-GUUAA--U ..((((....(((((((((...((....))..........(((....)))..))))))..((((.(........).)))).....----...)-))....)-)))..--. ( -26.70) >DroMoj_CAF1 251255 102 - 1 AAAAUUAUGGUCAGCGGCAGCAGCUGAAGCGUCAGACCCAGCCGCGAGGCAUUGCCGCAGUAGGUGAAAAGUUCCUCCUGAAAAAACAAACA--------UUCGUAAGAU ....((((((...(((((((...((((....)))).....(((....))).)))))))..((((.(........).))))............--------.))))))... ( -29.00) >DroPer_CAF1 231332 103 - 1 AAAAUUAUGGUCAGCGGCAGCAGCUGGAGCGUCAGACCCAGCCGCGAGGCAUUGCCGCAAUAGGUGUACAGUUCCUCCUAAGGAC----GAGG-GACAGAUGAGUAU--C ......(((.((((((((((....(((..(....)..)))(((....))).)))))))............(((((((........----))))-)))...))).)))--. ( -35.90) >consensus AAAAUUAUGGUCAGCGGCAGCAGCUGAAGCGUCAAACCCAGCCGCGAGGCAUUGCCGCAAUAGGUGAACAGUUCCUCCUAAAAAA____AAAG_GA____UUAGUAA__U .............((((((...((....))..........(((....)))..))))))..((((.(........).)))).............................. (-24.79 = -24.65 + -0.14)


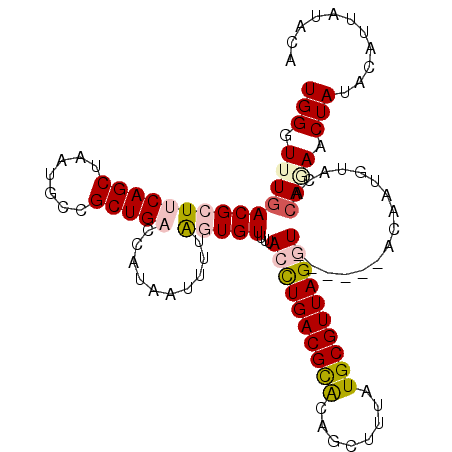
| Location | 16,235,778 – 16,235,883 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.48 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -17.95 |
| Energy contribution | -18.57 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16235778 105 + 22407834 UGGUUUUGACGCUUCAGCUAAUGCCGCUGACCAUAAUUUUAGUGUUUUACCUGACGCACAGCUUUAUGCGUUAGGU------ACAAUGUACACAGAACUAUACAUUAUACA ((((((((((((((((((.......)))))..........)))))..(((((((((((........))))))))))------).........))))))))........... ( -31.10) >DroPse_CAF1 222404 104 + 1 UGGGUCUGACGCUCCAGCUGCUGCCGCUGACCAUAAUUUUGAUGUUUUACUUGACGCGCUGCUUUAUGCGUUAGAUUAUACUAGUAUAUACACACAACUAUAU-----A-- ..((((((((((...(((.((.((.......((......))..(((......))))))).)))....)))))))))).......((((((........)))))-----)-- ( -21.80) >DroSec_CAF1 153110 105 + 1 UGGUUUUGACGCUUCAGCUAAUGCCGCUGACCAUAAUUUUAGUGUUUUACCUGACGCACAGCUUUAUGCGUUAGGU------ACAAUGUACACAGAACUAUACAUUAUACA ((((((((((((((((((.......)))))..........)))))..(((((((((((........))))))))))------).........))))))))........... ( -31.10) >DroSim_CAF1 155180 105 + 1 UGGUUUUGACGCUUCAGCUAAUGCCGCUGACCAUAAUUUUAGUGUUUUACCUGACGCACAGCUUUAUGCGUUAGGU------ACAAUGUACACAGAACUAUACAUUAUACA ((((((((((((((((((.......)))))..........)))))..(((((((((((........))))))))))------).........))))))))........... ( -31.10) >DroYak_CAF1 165753 104 + 1 UGGGUUUGACGCUUCAGCUGAUGCCGCUGACCAUAAUUUUAGUGUUUCACCUGACGUACACCUUUAUGCGUUAGGU------AUAAUGCACA-AAAACUAUACAUUAUAAA .(..((((.....(((((.......)))))...........(((((..((((((((((........))))))))))------..))))).))-))..)............. ( -25.90) >DroPer_CAF1 231395 104 + 1 UGGGUCUGACGCUCCAGCUGCUGCCGCUGACCAUAAUUUUGAUGUUUUACUUGACGCGCUGCUUUAUGCGUUAGAUUAUACUAGUAUAUACACACAACUAUAU-----A-- ..((((((((((...(((.((.((.......((......))..(((......))))))).)))....)))))))))).......((((((........)))))-----)-- ( -21.80) >consensus UGGGUUUGACGCUUCAGCUAAUGCCGCUGACCAUAAUUUUAGUGUUUUACCUGACGCACAGCUUUAUGCGUUAGGU______ACAAUGUACACAGAACUAUACAUUAUACA (((.((((((((((((((.......)))))..........)))))...((((((((((........))))))))))................)))).)))........... (-17.95 = -18.57 + 0.61)


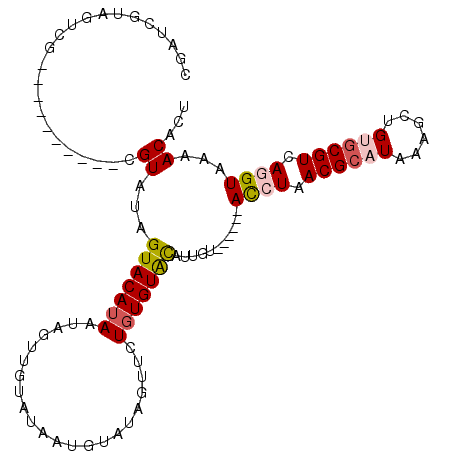
| Location | 16,235,818 – 16,235,913 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.32 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -12.35 |
| Energy contribution | -12.77 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16235818 95 - 22407834 CGAUCGUAGUCG----------CGUAUAGUACAUAAUAGUUGUAUAAUGUAUAGUUCUGUGUACAUUGU------ACCUAACGCAUAAAGCUGUGCGUCAGGUAAAACACU ((((....))))----------......(((((.......)))))((((((((......)))))))).(------((((.(((((((....))))))).)))))....... ( -24.20) >DroPse_CAF1 222444 103 - 1 CCAUCAACGUCGUUCGCAUACUCGUAUAGUACAUAAUUG---U-----AUAUAGUUGUGUGUAUAUACUAGUAUAAUCUAACGCAUAAAGCAGCGCGUCAAGUAAAACAUC ......(((.((((.(((((((.(((((..(((((((((---.-----...)))))))))...))))).)))))...(....)......)))))))))............. ( -21.60) >DroSec_CAF1 153150 95 - 1 CGAUCGUAGUCG----------CGUAUAGUACAUAAUAGUUGUAUAAUGUAUAGUUCUGUGUACAUUGU------ACCUAACGCAUAAAGCUGUGCGUCAGGUAAAACACU ((((....))))----------......(((((.......)))))((((((((......)))))))).(------((((.(((((((....))))))).)))))....... ( -24.20) >DroSim_CAF1 155220 95 - 1 CGAUCGUAGUCG----------CGUAUAGUACAUAAUAGUUGUAUAAUGUAUAGUUCUGUGUACAUUGU------ACCUAACGCAUAAAGCUGUGCGUCAGGUAAAACACU ((((....))))----------......(((((.......)))))((((((((......)))))))).(------((((.(((((((....))))))).)))))....... ( -24.20) >DroYak_CAF1 165793 94 - 1 CGAUCGUAGUCG----------CGUAUAGUACAUAAUAGUUUUAUAAUGUAUAGUUUU-UGUGCAUUAU------ACCUAACGCAUAAAGGUGUACGUCAGGUGAAACACU ((((....))))----------................(((((((.(((((((.((((-(((((.(((.------...))).))))))))))))))))...)))))))... ( -20.70) >DroPer_CAF1 231435 103 - 1 CCAUCAACGUCGUUCGCAUACUCGUAUAGUACAUAAUUG---U-----AUAUAGUUGUGUGUAUAUACUAGUAUAAUCUAACGCAUAAAGCAGCGCGUCAAGUAAAACAUC ......(((.((((.(((((((.(((((..(((((((((---.-----...)))))))))...))))).)))))...(....)......)))))))))............. ( -21.60) >consensus CGAUCGUAGUCG__________CGUAUAGUACAUAAUAGUUGUAUAAUGUAUAGUUCUGUGUACAUUGU______ACCUAACGCAUAAAGCUGUGCGUCAGGUAAAACACU .......................((...(((((((......................)))))))...........((((.((((((......)))))).))))...))... (-12.35 = -12.77 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:42 2006