
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,233,320 – 16,233,416 |
| Length | 96 |
| Max. P | 0.641385 |

| Location | 16,233,320 – 16,233,416 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.52 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -16.67 |
| Energy contribution | -16.92 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16233320 96 + 22407834 GUGGUGUCCAACAAUGGAUAUGUUCUGCUGCAUCCUGAUCUACGACCCAUUGGGGUAAGUUGAAUGGUGUCAAACAGUGU-UUACA-------UAUAUAUAUAU ...(((((((....)))))))((((.(((..((((..((.........))..)))).))).)))).((((.((((...))-)))))-------).......... ( -20.00) >DroVir_CAF1 216513 102 + 1 GUGGUUUCCAACAAUGGCUACAUGCUGCUGCAUCCGGAUCUGCGACCCAUUGGCGUAAGCAAGAUACUUUUAACCAGCCAGAAAAUUGU-AGUUACCU-AUUAU ((((((.........))))))((((....))))..((..((((((....(((((......................)))))....))))-))...)).-..... ( -19.35) >DroSec_CAF1 150640 92 + 1 GUGGUGUCCAACAAUGGAUAUGUUCUGCUGCAUCCCGAUCUGCGACCCAUUGGGGUAAGUUGAUUGGUGUCAAAUGGUGU-UGAUA-------UAU----AUAU (..(((((((....))))).......))..)...((((((.((.((((....))))..)).))))))((((((......)-)))))-------...----.... ( -23.81) >DroSim_CAF1 152720 96 + 1 GUGGUGUCCAACAAUGGAUAUGUUCUGCUUCAUCCUGAUCUACGACCCAUUGGGGUAAGUUGAAUGGUAUCAAAUGGUGU-UGAUA-------UAUAUAUAUAU ...(((((((....)))))))((((.((((.((((..((.........))..)))))))).)))).(((((((......)-)))))-------).......... ( -23.10) >DroEre_CAF1 154333 94 + 1 GUGGUGUCCAACAAUGGAUAUGUUCUGCUGCAUCCCGAUCUACGACCCAUUGGGGUAAGCUGAACCAUAUCAACCGGGGC-AUAUG---------UAUAUGUAU ...((((((.......(((((((((.(((..((((((((.........)))))))).))).))).)))))).....))))-))...---------......... ( -29.60) >DroYak_CAF1 163100 95 + 1 GUGGUGUCCAACAAUGGAUAUGUUCUGCUGCAUCCCGAUCUACGACCCAUUGGGGUAAGCUGAACCAUAUCACC-------AUAUG-GUGAA-UUCUUAUACAU ((((((((((....)))))))((((.(((..((((((((.........)))))))).))).))))))).(((((-------....)-)))).-........... ( -30.70) >consensus GUGGUGUCCAACAAUGGAUAUGUUCUGCUGCAUCCCGAUCUACGACCCAUUGGGGUAAGCUGAAUGGUAUCAAACGGUGU_AUAUA_______UAUAUAUAUAU ...(((((((....)))))))((((.(((..((((((((.........)))))))).))).))))....................................... (-16.67 = -16.92 + 0.25)

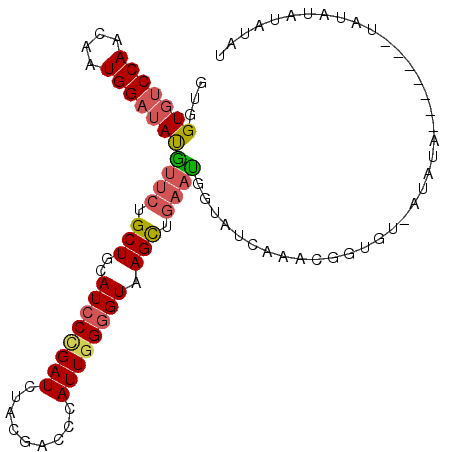
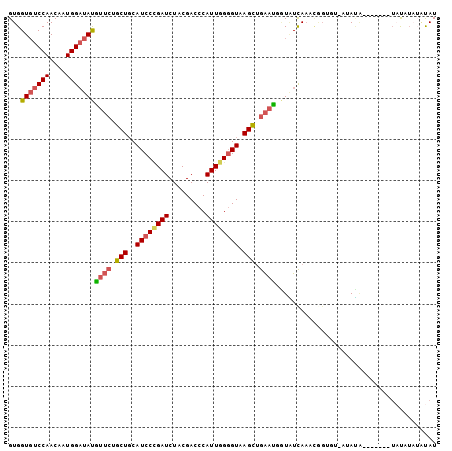
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:39 2006