| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,231,224 – 16,231,314 |
| Length | 90 |
| Max. P | 0.725534 |

| Location | 16,231,224 – 16,231,314 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 71.96 |
| Mean single sequence MFE | -16.27 |
| Consensus MFE | -1.10 |
| Energy contribution | -1.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.07 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16231224 90 + 22407834 CAUGUUCUUUUGCCAAA------GAAAAAAACAAGUUUUGUAGAGGGAUACCCAUACCAAUUAAGUACAAAAGUGGUCCUUACAAAUUCAAUCGCU ....((((((....)))------)))..........(((((((.(((...)))..((((.((........)).))))..))))))).......... ( -10.60) >DroSec_CAF1 148679 83 + 1 CAUGUUCAAUGCCAGAA------AGAAAAAACAAGUUGUGUGAAGGGAUAACACAACCAAUUAAGUACAAAAGU-------ACAAUUUCAAUGGCU ..........((((...------.((((......(((((((.........))))))).......((((....))-------))..))))..)))). ( -18.80) >DroSim_CAF1 150795 83 + 1 CAUGUUCAAUGCCAGAA------AGAAAAAACAAGUUGUGUGAAGGGAUAACACAACCAAUUAAGUACAAAAGU-------ACAAUUUCAAUGGCU ..........((((...------.((((......(((((((.........))))))).......((((....))-------))..))))..)))). ( -18.80) >DroEre_CAF1 152202 96 + 1 CAUGUUUGGCUGCAAAAAAAUUGCAAAGGGAAAAAUUUCGUAAAGGGAUGACCCCACCAAUUAAGUAAAAAUAUGGGCCUUACAAUUUCAAUGACU ..(((..((((((((.....))))...(((....((..(.....)..))...)))....................))))..)))............ ( -19.60) >DroYak_CAF1 160327 89 + 1 CAAGUUUGUAGGCAU-------ACAAAAAAACAAGUUUUGUGAAGGGAUAACCCUACCAACUAAGUACAAAAAUGGGCUUUACACUUUUAAUGACU .(((..(((((((.(-------((((((.......))))))).((((....)))).(((..............))))).)))))..)))....... ( -13.54) >consensus CAUGUUCAAUGGCAAAA______AAAAAAAACAAGUUUUGUGAAGGGAUAACCCAACCAAUUAAGUACAAAAGUGG_C_UUACAAUUUCAAUGGCU ............................................((..........))...................................... ( -1.10 = -1.10 + -0.00)

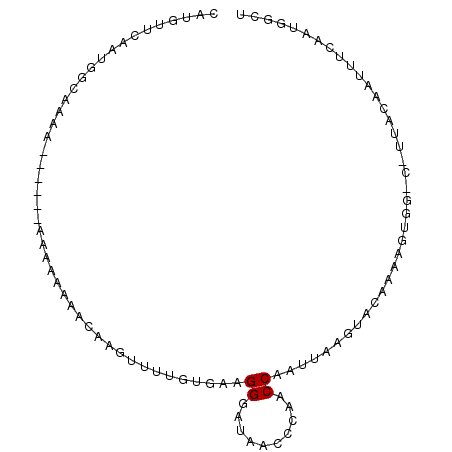
| Location | 16,231,224 – 16,231,314 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 71.96 |
| Mean single sequence MFE | -17.40 |
| Consensus MFE | -0.14 |
| Energy contribution | -0.90 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.01 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16231224 90 - 22407834 AGCGAUUGAAUUUGUAAGGACCACUUUUGUACUUAAUUGGUAUGGGUAUCCCUCUACAAAACUUGUUUUUUUC------UUUGGCAAAAGAACAUG ..........(((((((((...((((..(((((.....)))))))))...))).))))))...((((((((((------....).))))))))).. ( -15.90) >DroSec_CAF1 148679 83 - 1 AGCCAUUGAAAUUGU-------ACUUUUGUACUUAAUUGGUUGUGUUAUCCCUUCACACAACUUGUUUUUUCU------UUCUGGCAUUGAACAUG .((((..((((..((-------((....))))..(((.((((((((.(.....).)))))))).))).)))).------...)))).......... ( -18.90) >DroSim_CAF1 150795 83 - 1 AGCCAUUGAAAUUGU-------ACUUUUGUACUUAAUUGGUUGUGUUAUCCCUUCACACAACUUGUUUUUUCU------UUCUGGCAUUGAACAUG .((((..((((..((-------((....))))..(((.((((((((.(.....).)))))))).))).)))).------...)))).......... ( -18.90) >DroEre_CAF1 152202 96 - 1 AGUCAUUGAAAUUGUAAGGCCCAUAUUUUUACUUAAUUGGUGGGGUCAUCCCUUUACGAAAUUUUUCCCUUUGCAAUUUUUUUGCAGCCAAACAUG .(.((..(((((((((((((((((...((.....))...))))).............(((....))).))))))))))))..)))........... ( -18.40) >DroYak_CAF1 160327 89 - 1 AGUCAUUAAAAGUGUAAAGCCCAUUUUUGUACUUAGUUGGUAGGGUUAUCCCUUCACAAAACUUGUUUUUUUGU-------AUGCCUACAAACUUG (((...((((((((.......)))))))).)))..((.(((((((....)))...((((((.......))))))-------.)))).))....... ( -14.90) >consensus AGCCAUUGAAAUUGUAA_G_CCACUUUUGUACUUAAUUGGUAGGGUUAUCCCUUCACAAAACUUGUUUUUUUG______UUCUGCCAACGAACAUG .....................................((..((((....))))...))...................................... ( -0.14 = -0.90 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:37 2006