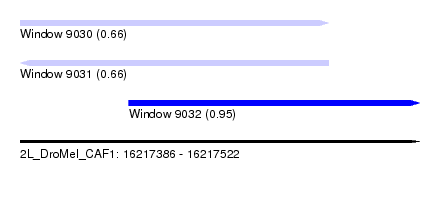
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,217,386 – 16,217,522 |
| Length | 136 |
| Max. P | 0.947967 |

| Location | 16,217,386 – 16,217,491 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 91.46 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.45 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.659915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16217386 105 + 22407834 AAAUCUCGGAGAUUUGAAAUCUCAUUGACAAUUGGAAAUGUGACCAAAAUCAUGCAUUUCCGUCGCCAUCUCUGGCCAAACCAGACCAAUGCAGCCAGCCGCCCA ...((...(((((.....)))))...)).....((((((((............))))))))((.((....(((((.....))))).....)).)).......... ( -21.80) >DroSec_CAF1 134728 98 + 1 AAAUCUCGGAGAUUUGAAAUCUCAUUGACAAUUGGAAAUGUGACCAAAAUCAUGCAUUUCCGUCGCCAUCUCUGGUCAAACCAGACCAGUGCAG-------CCCA ........(((((.....)))))...(((....((((((((............)))))))))))((...(.((((((......)))))).)..)-------)... ( -25.00) >DroSim_CAF1 136142 98 + 1 AAAUCUCGGAGAUUUGAAAUCUCAUUGACAAUUGGAAAUGUGACCAAAAUCAUGCAUUUCCGUCGCCAUCUCUGGUCAAACCAGACCAGUGCAG-------CCCA ........(((((.....)))))...(((....((((((((............)))))))))))((...(.((((((......)))))).)..)-------)... ( -25.00) >DroEre_CAF1 138282 98 + 1 AAAUCUCGGAGAUUUGAAAUCUCAUUGACAAUUGGAAAUGUGACCAAAAUCAUGCAUUUCCGUCGCCAUCUCUGACCAGAGUAGACCAGACCAG-------GGCA ........(((((.....)))))...(((....((((((((............)))))))))))(((...((((..(......)..))))....-------))). ( -22.50) >consensus AAAUCUCGGAGAUUUGAAAUCUCAUUGACAAUUGGAAAUGUGACCAAAAUCAUGCAUUUCCGUCGCCAUCUCUGGCCAAACCAGACCAGUGCAG_______CCCA ........(((((.....)))))...(((....((((((((............)))))))))))((....(((((.....))))).....))............. (-19.14 = -19.45 + 0.31)

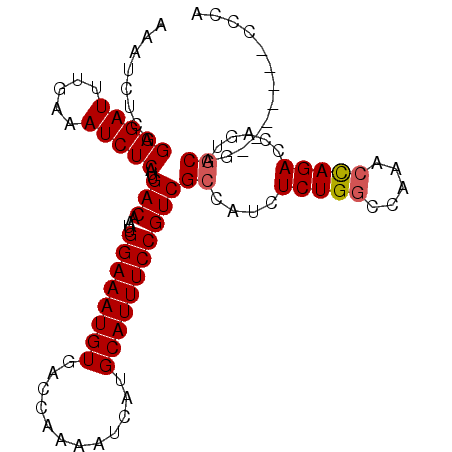
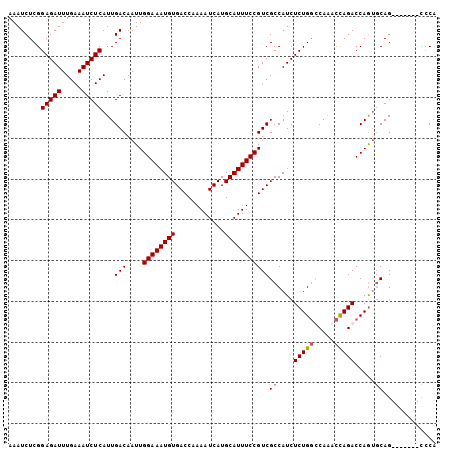
| Location | 16,217,386 – 16,217,491 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 91.46 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -26.60 |
| Energy contribution | -25.98 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16217386 105 - 22407834 UGGGCGGCUGGCUGCAUUGGUCUGGUUUGGCCAGAGAUGGCGACGGAAAUGCAUGAUUUUGGUCACAUUUCCAAUUGUCAAUGAGAUUUCAAAUCUCCGAGAUUU ...((((....)))).((((...((((((((((....))))((((((((((..((((....)))))))))))....)))..........)))))).))))..... ( -31.60) >DroSec_CAF1 134728 98 - 1 UGGG-------CUGCACUGGUCUGGUUUGACCAGAGAUGGCGACGGAAAUGCAUGAUUUUGGUCACAUUUCCAAUUGUCAAUGAGAUUUCAAAUCUCCGAGAUUU ...(-------((((.((((((......)))))).).))))((((((((((..((((....)))))))))))....)))...(((((.....)))))........ ( -28.70) >DroSim_CAF1 136142 98 - 1 UGGG-------CUGCACUGGUCUGGUUUGACCAGAGAUGGCGACGGAAAUGCAUGAUUUUGGUCACAUUUCCAAUUGUCAAUGAGAUUUCAAAUCUCCGAGAUUU ...(-------((((.((((((......)))))).).))))((((((((((..((((....)))))))))))....)))...(((((.....)))))........ ( -28.70) >DroEre_CAF1 138282 98 - 1 UGCC-------CUGGUCUGGUCUACUCUGGUCAGAGAUGGCGACGGAAAUGCAUGAUUUUGGUCACAUUUCCAAUUGUCAAUGAGAUUUCAAAUCUCCGAGAUUU .(((-------....((((..(......)..))))...)))((((((((((..((((....)))))))))))....)))...(((((.....)))))........ ( -27.10) >consensus UGGG_______CUGCACUGGUCUGGUUUGACCAGAGAUGGCGACGGAAAUGCAUGAUUUUGGUCACAUUUCCAAUUGUCAAUGAGAUUUCAAAUCUCCGAGAUUU ................((((((......))))))...((((((.(((((((..((((....)))))))))))..))))))..(((((.....)))))........ (-26.60 = -25.98 + -0.63)

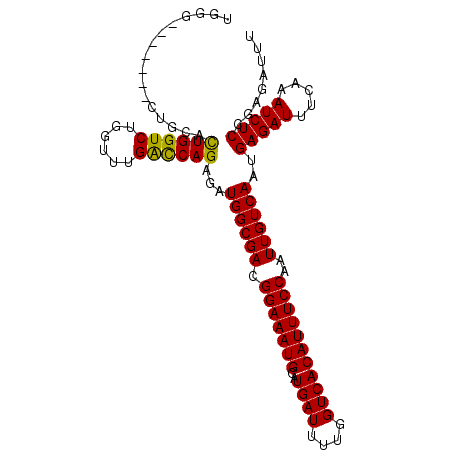
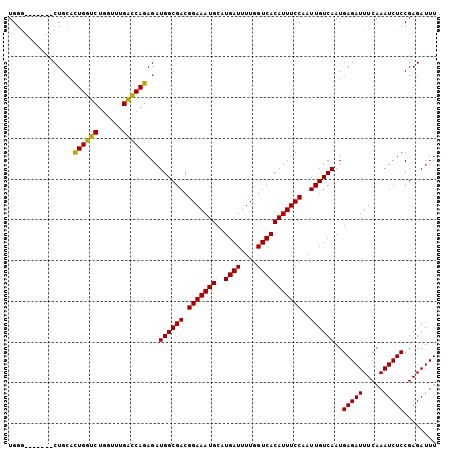
| Location | 16,217,423 – 16,217,522 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.73 |
| Mean single sequence MFE | -20.61 |
| Consensus MFE | -10.75 |
| Energy contribution | -12.31 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16217423 99 + 22407834 AUGUGACCAAAAUCAUGCAUUUCCGUCGCCAUCUCUGGCCAAACCAGACCAAUGCAGCCAGCCGCCCAUCGCUGCCCACCCCCA---AAGCC------CAAAUCCACC ..(((((..((((.....))))..)))))....(((((.....))))).....(((((.(........).))))).........---.....------.......... ( -16.60) >DroSec_CAF1 134765 91 + 1 AUGUGACCAAAAUCAUGCAUUUCCGUCGCCAUCUCUGGUCAAACCAGACCAGUGCAG-------CCCAUCGCUGCACAAAACCA---AA-CC------CAAACCCACC ..(((((..((((.....))))..)))))......(((((......)))))((((((-------(.....))))))).......---..-..------.......... ( -23.90) >DroSim_CAF1 136179 91 + 1 AUGUGACCAAAAUCAUGCAUUUCCGUCGCCAUCUCUGGUCAAACCAGACCAGUGCAG-------CCCAUCGCUGCACAAAACCA---AA-CC------CAAGCCCACC ..(((((..((((.....))))..)))))......(((((......)))))((((((-------(.....))))))).......---..-..------.......... ( -23.90) >DroEre_CAF1 138319 99 + 1 AUGUGACCAAAAUCAUGCAUUUCCGUCGCCAUCUCUGACCAGAGUAGACCAGACCAG-------GGCAAUGCAACCCAGCCC-AGGCAA-CCAACCAACCAGCCGACC ............((.((((((((((((.....((((....)))).......)))..)-------)).)))))).........-.(((..-...........))))).. ( -18.02) >consensus AUGUGACCAAAAUCAUGCAUUUCCGUCGCCAUCUCUGGCCAAACCAGACCAGUGCAG_______CCCAUCGCUGCACAAAACCA___AA_CC______CAAACCCACC ..(((((..((((.....))))..)))))....(((((.....)))))...((((((..............))))))............................... (-10.75 = -12.31 + 1.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:32 2006