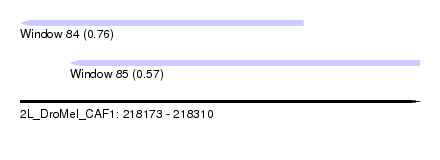
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 218,173 – 218,310 |
| Length | 137 |
| Max. P | 0.762366 |

| Location | 218,173 – 218,270 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 84.74 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -21.40 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
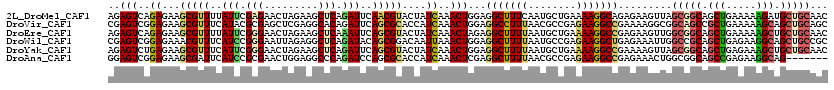
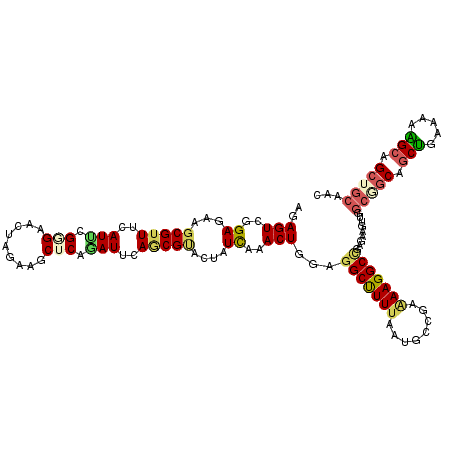
>2L_DroMel_CAF1 218173 97 - 22407834 AUUCAACGUACUAUCAAACUGGAGGCUUUCAAUGCUGAAAAGGCAGAGAAGUUAGCGGCAGCUGAAAAAGAUGCUGCAACUGAAAAAGCUGCGGCGA ......(((.........((...(((.......)))....))((((.....((((..(((((..........)))))..)))).....)))).))). ( -26.00) >DroSec_CAF1 10472 97 - 1 AUUCAACGUACUAUCAAACUGGAGGCUUUUAAUGCUGAAAAGGCGGAGAAGUUAGCGGCAGCUGAAAUAGCUGCUGCAACUGAAAAAGCUGCGGCGA ..................(((..(((((((..((((.....))))....((((.((((((((((...))))))))))))))..))))))).)))... ( -30.50) >DroSim_CAF1 10464 97 - 1 AUUCAACGUACUAUCAAACUGGAGGCUUUUAAUGCUGAAAAGGCGGAGAAGUUAGCGGCAGCUGAAAAAGCUGCUGCAACUGAAAAAGCUGCGGCGA ..................(((..(((((((..((((.....))))....((((.(((((((((.....)))))))))))))..))))))).)))... ( -31.50) >DroEre_CAF1 10763 97 - 1 AUUCAGCGUACUAUCAAACUAGAGGCUUUUAAUGCUGAAAAGGCCGAGAAGUUGGCGGCAGCUGAAAAAGCUGCUGCAACUGAGAAAGCUGCGGCAA .((((((((....((......))........))))))))...((((((.(((((.((((((((.....))))))))))))).......)).)))).. ( -34.80) >DroWil_CAF1 12421 81 - 1 AUACAGCGGACAAUUAAACUGGAGGCUUUUAAUGCCGAGAAGGCUGAGAAAUUGGCCGCAGCUGAGAAGGCAGCUGCCGCU---------------- ....((((...............(((.......))).....(((..(....)..)))(((((((......)))))))))))---------------- ( -30.80) >DroYak_CAF1 12519 97 - 1 AUUCAGCGUACUAUCAAACUGGAGGCUUUUAAUGCUGAAAAGGCCGAAAAGUUAGCGGCAGCUGAGAAAGCUGCUGCAACGGAAAAAGCUGCUGCAA ...((((.........((((...(((((((........)))))))....)))).(((((((((.....)))))))))..........))))...... ( -33.70) >consensus AUUCAACGUACUAUCAAACUGGAGGCUUUUAAUGCUGAAAAGGCCGAGAAGUUAGCGGCAGCUGAAAAAGCUGCUGCAACUGAAAAAGCUGCGGCGA .......................(((((((...(((.....)))..))))))).(((((((((.....)))))))))..........((....)).. (-21.40 = -21.90 + 0.50)

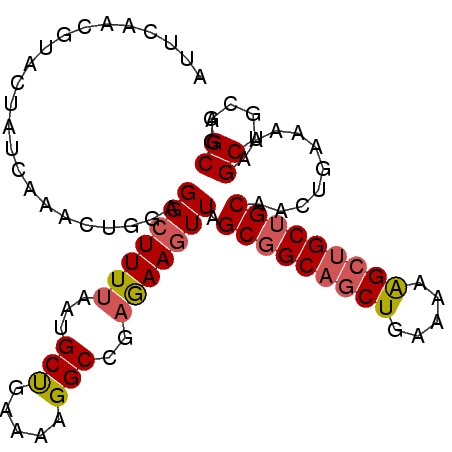
| Location | 218,190 – 218,310 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -22.95 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 218190 120 - 22407834 AGAGUCAGAGAAGCGUUUUAUUCGAGAACUAGAAGCUCAGAUUCAACGUACUAUCAAACUGGAGGCUUUCAAUGCUGAAAAGGCAGAGAAGUUAGCGGCAGCUGAAAAAGAUGCUGCAAC .(((((.(((....(((((.....)))))......))).)))))...................((((((...((((.....))))..)))))).((((((.((.....)).))))))... ( -28.80) >DroVir_CAF1 13431 120 - 1 CGAGUCGGAGAAGCGUUUCAUACGCGAGCUCGAGGCACAGAUUCAGCGCACCAUCAAACUGGAGGCCUUUAACGCCGAGAAGGCCGAAAAGGCGGCAGCCGCUGAAAAAGCAGCUGCAGC .(((((.(((..((((.....))))...)))(.....).)))))..(((.(((......))).(((((((........)))))))......)))(((((.(((.....))).)))))... ( -43.90) >DroEre_CAF1 10780 120 - 1 AGAGUCAGAGAAGCGUUUUAUUCGGGAACUAGAAGCUCAAAUUCAGCGUACUAUCAAACUAGAGGCUUUUAAUGCUGAAAAGGCCGAGAAGUUGGCGGCAGCUGAAAAAGCUGCUGCAAC .((((....(((........)))(....).....)))).....((((...(((......))).(((((((........))))))).....))))(((((((((.....)))))))))... ( -34.00) >DroWil_CAF1 12422 120 - 1 CGAGUCGGAGAAACGUUUCAUCCGGGAAUUAGAGGCUCAGAUACAGCGGACAAUUAAACUGGAGGCUUUUAAUGCCGAGAAGGCUGAGAAAUUGGCCGCAGCUGAGAAGGCAGCUGCCGC .(((.((......)).))).(((((..(((((((((((((..................)))..)))))))))).))).)).(((..(....)..)))(((((((......)))))))... ( -39.97) >DroYak_CAF1 12536 120 - 1 AGAGUCUGAGAAGCGUUUCAUUCGGGAACUAGAAGCUCAGAUUCAGCGUACUAUCAAACUGGAGGCUUUUAAUGCUGAAAAGGCCGAAAAGUUAGCGGCAGCUGAGAAAGCUGCUGCAAC .(((((((((....(((((.....)))))......)))))))))............((((...(((((((........)))))))....)))).(((((((((.....)))))))))... ( -45.00) >DroAna_CAF1 13477 113 - 1 GGAGUCGGAGAAGCGAUUCAUCCGCGAACUGGAGGCCCAGAUCCAGCGCACCAUCAAACUCGAGGCUUUUAACGCCGAGAAGGCCGAGAAACUGGCGGCAGCCGAGAAGGCAG------- .((((((......))))))..(((((..(((((........)))))..)..........((..(((((((........)))))))..)).....))))..(((.....)))..------- ( -36.30) >consensus AGAGUCGGAGAAGCGUUUCAUUCGGGAACUAGAAGCUCAGAUUCAGCGUACUAUCAAACUGGAGGCUUUUAAUGCCGAAAAGGCCGAGAAGUUGGCGGCAGCUGAAAAAGCAGCUGCAAC ..(((..((...(((((..(((.(((.........))).)))..)))))....))..)))...(((((((........))))))).........(((((.(((.....))).)))))... (-22.95 = -23.48 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:32 2006