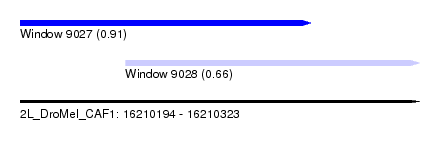
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,210,194 – 16,210,323 |
| Length | 129 |
| Max. P | 0.908478 |

| Location | 16,210,194 – 16,210,288 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.16 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -16.03 |
| Energy contribution | -16.03 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
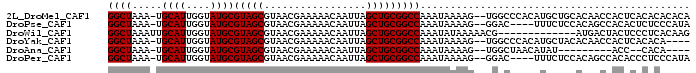
>2L_DroMel_CAF1 16210194 94 + 22407834 GGCUAAA-UGCAUUGGUAUGCGUAGCGUAACGAAAAACAAUUAGCUGCGGCCAAAUAAAAG--UGGCCCACAUGCUGCACAACCACUCACACACACA .((....-.))..((((.((.(((((((...(.....).....(.((.(((((........--))))))))))))))).))))))............ ( -22.80) >DroPse_CAF1 183736 90 + 1 GGCUAAA-UGCAUUGGUAUGCGUAGCGUAACGAAAAACAAUUAGCUGCGGCCAAAUAAAAG--GGAC----UUUCUCCACAGCCACACUCUCCCAUA ((((...-....(((((...((((((.................))))))))))).......--(((.----....)))..))))............. ( -20.63) >DroWil_CAF1 187921 84 + 1 GGCUAAAUUGCAUUGGUAUGCGUAGCGUAACGAAAAACAAUUAGCUGCGGCCAAAUAUAAAAACG-------------AUGACUACUCCCUCACAAG ((((.....((((....))))(((((.................))))))))).............-------------................... ( -16.03) >DroYak_CAF1 137996 90 + 1 GGCUAAA-UGCAUUGGUAUGCGUAGCGUAACGAAAAACAAUUAGCUGCGGCCAAAUAAAAG--UGGCCCACAUGCUACACAACCACUCACACA---- .((....-.))..((((.((.(((((((...(.....).....(.((.(((((........--))))))))))))))).))))))........---- ( -22.80) >DroAna_CAF1 123208 79 + 1 GGCUAAA-UGCAUUGGUAUGCGUAGCGUAACGAAAAACAAUUAGCUGCGGCCAAAUAAAAG--UGGCUAACAUAU---------ACC--CACA---- ((((...-.((((....))))(((((.................)))))))))........(--(((.((.....)---------).)--))).---- ( -20.43) >DroPer_CAF1 194076 90 + 1 GGCUAAA-UGCAUUGGUAUGCGUAGCGUAACGAAAAACAAUUAGCUGCGGCCAAAUAAAAG--GGAC----UUUCUCCACAGCCACACCCUCCCAUA ((((...-....(((((...((((((.................))))))))))).......--(((.----....)))..))))............. ( -20.63) >consensus GGCUAAA_UGCAUUGGUAUGCGUAGCGUAACGAAAAACAAUUAGCUGCGGCCAAAUAAAAG__UGGC____AUGCU_CACAACCACUCACACACA_A ((((.....((((....))))(((((.................)))))))))............................................. (-16.03 = -16.03 + -0.00)

| Location | 16,210,228 – 16,210,323 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.37 |
| Mean single sequence MFE | -15.68 |
| Consensus MFE | -11.92 |
| Energy contribution | -11.76 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16210228 95 + 22407834 AACAAUUAGCUGCGGCCAAAUAAAAGUGGCCCACAUGCUGCACAACCACUCACACACACACACAAUGUCUUUGCACUCGGCAAAA---UGAUAC--AGCU .......(((((.(((((........)))))..................................(((((((((.....))))).---.)))))--)))) ( -18.50) >DroSec_CAF1 127665 84 + 1 AACAAUUAGCUGCGGCCAAAUAAAAGUGGCCCUCAUGCUGCACAACCACUCACACACACACACAAUGUCUUUGCACUCGGCAAA---------------- ......((((((.(((((........)))))..)).))))..............................((((.....)))).---------------- ( -14.40) >DroSim_CAF1 129159 84 + 1 AACAAUUAGCUGCGGCCAAAUAAAAGUGGCCCACAUGCUGCACAACCACUCACACACAUACACAAUGUCUUUGCACUCGGCAAA---------------- ......((((((.(((((........)))))..)).))))...............((((.....))))..((((.....)))).---------------- ( -14.60) >DroEre_CAF1 131212 96 + 1 AACAAUUAGCUGCGGCCAAAUAAAAGUGGCCCACAUGCUGCACAGCCACUCACACA----CACAUUGUCUCUGCACUCAAGAAAAAUUUGACAUAAACCU ..(((..(((((.(((((........)))))..)).)))(((.((.((........----.....)).)).))).............))).......... ( -13.92) >DroYak_CAF1 138030 94 + 1 AACAAUUAGCUGCGGCCAAAUAAAAGUGGCCCACAUGCUACACAACCACUCACACA----CACAUUGUCCGUGCACUCGGCAAAAAUUUAACAA--AGCC ......((((((.(((((........)))))..)).))))................----....(((.(((......))))))...........--.... ( -17.00) >consensus AACAAUUAGCUGCGGCCAAAUAAAAGUGGCCCACAUGCUGCACAACCACUCACACACA_ACACAAUGUCUUUGCACUCGGCAAAA___U_A_A___A_C_ ......((((((.(((((........)))))..)).))))............................................................ (-11.92 = -11.76 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:29 2006