| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,202,678 – 16,202,777 |
| Length | 99 |
| Max. P | 0.802470 |

| Location | 16,202,678 – 16,202,777 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
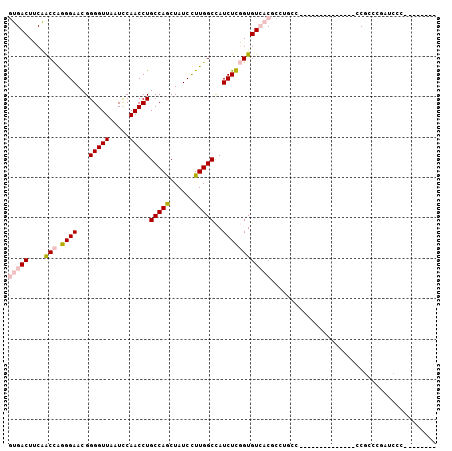
| Mean pairwise identity | 72.30 |
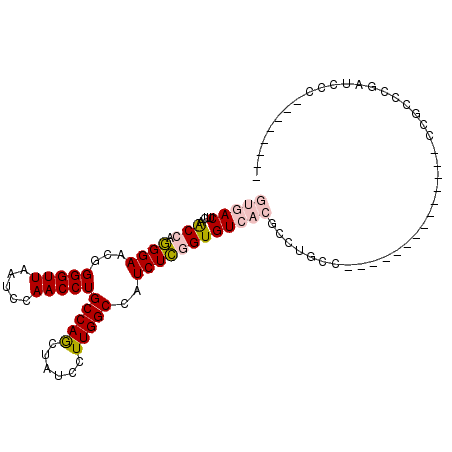
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.57 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802470 |
| Prediction | RNA |
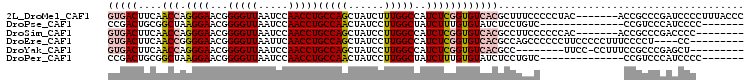
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16202678 99 + 22407834 GUGACUUCAACCAGGGAACGGGGUUAAUCCAACCUGCCAGCUAUCUUUGGCCAUCUCGGUGUCACGCUUUCCCCCUAC-------ACCGCCCGAUCCCCUUUACCC .............((((.((((.............(((((......))))).....((((((..............))-------)))))))).))))........ ( -25.64) >DroPse_CAF1 176697 85 + 1 CCGACUGCGGCUAAGGAACGGGGUUAAUCCAACCUGCCAACUAUCCUUGGCUAUCUUUGUGUAUCUCCUGUC--------------CCGUCCCAUCCCC------- ......((((...((((...(((((.....)))))(((((......)))))..............))))...--------------)))).........------- ( -19.50) >DroSim_CAF1 121685 91 + 1 GUGACUUCAACCAGGGAACGGGGUUAAUCCAACCUGCCAGCUAUCCUUGGCCAUCUCGGUGUCACGCCUUCCCCCCAC-------ACCGCCCGACCCC-------- ((..(((.....)))..))((((((..........(((((......))))).....((((((..............))-------))))...))))))-------- ( -22.54) >DroEre_CAF1 123800 93 + 1 GUGACUUCAACCGGGGAACGGGGUUAAUUCAACCUGCCAGCUAUCCUUGGCCAUCUCGGUGUCACGCCAGCCCCCCUUCCCCCUUUCCCCU----CC--------- ............((((((.((((((..........(((((......)))))......((((...))))))))))..)))))).........----..--------- ( -28.40) >DroYak_CAF1 130501 88 + 1 GUGACUUCAACCAGGGAACGGGGUUAAUCCAACCUGCCAGCUAUCCUUGGCCAUCUCGGUGUCACGCC--------UUCC-CCUUUCCGCCCGAGCU--------- (((((....(((.((((...(((((.....)))))(((((......)))))..))))))))))))...--------....-.......((....)).--------- ( -21.70) >DroPer_CAF1 186952 85 + 1 CCGACUGCGGCUAAGGAACGGGGUUAAUCCAACCUGCCAACUAUCCUUGGCUAUCUUUGUGUAUCUCCUGUC--------------CCGUCCCAUCCCC------- ......((((...((((...(((((.....)))))(((((......)))))..............))))...--------------)))).........------- ( -19.50) >consensus GUGACUUCAACCAGGGAACGGGGUUAAUCCAACCUGCCAGCUAUCCUUGGCCAUCUCGGUGUCACGCCUGCC______________CCGCCCGAUCCC________ (((((....(((.((((...(((((.....)))))(((((......)))))..))))))))))))......................................... (-16.12 = -16.57 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:24 2006