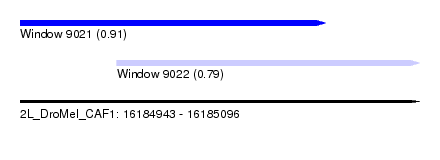
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,184,943 – 16,185,096 |
| Length | 153 |
| Max. P | 0.913720 |

| Location | 16,184,943 – 16,185,060 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -27.15 |
| Energy contribution | -26.52 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
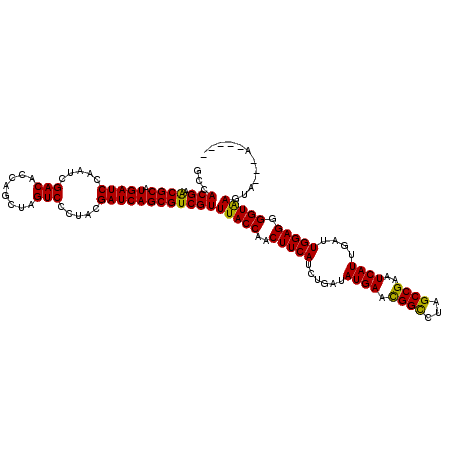
>2L_DroMel_CAF1 16184943 117 + 22407834 ---AAACGUGCAGUCUACGCACCCAAUGGUGCUCUUCCUGGCCACGAACGCAUGAUCCAAUCGACACCAGCUAGUCCCUACGAUCAGCGUCGUUUACCAACUUCAUCUGAUAUGAACGGU ---...(((.(((((...(((((....)))))......(((..((((.(((.(((((.....(((........))).....))))))))))))...))).........))).)).))).. ( -30.60) >DroVir_CAF1 150978 117 + 1 ---AUUUAUAUAGUCUACGCGCCCAAUGGUGCUAUUCCUGGCCACGAACGCAUGAUCCAAUCGACACCAGCUAGUCCCUACGAUCAGCGUCGUUUACCAACUUCAUCUGAAAUGAACGGC ---...............(((((....)))))........(((((((.(((.(((((.....(((........))).....))))))))))))........(((((.....))))).))) ( -28.00) >DroGri_CAF1 113172 117 + 1 ---AAUUAUAUAGUCUACGCACCCAAUGGUGCUAUUCCUGGCCACGAACGCAUGAUCCAAUCGACACCAGCUAGUCCUUACGAUCAGCGUCGUUUACCAACUUCAUCUGAAAUGAAUGGC ---...............(((((....)))))........(((((((.(((.(((((.....(((........))).....))))))))))))........(((((.....))))).))) ( -28.20) >DroWil_CAF1 146839 117 + 1 ---AUACCAUUAGUCUACGCACCCAAUGGUGCUAUUCCUGGCCACGAGCGCAUGAUCCAAUCGACACCAGCUAGUCCUUACGAUCAGCGUCGUUUACCAACUUCAUCUGAUAUGAACGGU ---..(((..........(((((....)))))......(((..((((.(((.(((((.....(((........))).....))))))))))))...)))..(((((.....))))).))) ( -29.20) >DroMoj_CAF1 170141 120 + 1 UUCAUACAUAUAGUCUACGCGGCCAAUGGUGCUAUUCCUGGCCACGAACGCAUGAUCCAAUCGACACCAGCUAGUCCCUACGAUCAGCGUCGUUUACCAACUUCAUCUGAUAUGAAUGGC .((((.(((((.........(((((..((.......)))))))((((.(((.(((((.....(((........))).....))))))))))))................))))).)))). ( -30.50) >DroPer_CAF1 138893 117 + 1 ---GCUCAUUUAGUCUACGCACCCAAUGGUGCUAUUCCUGGCCACGAGCGCAUGAUCCAAUCGACCCCAGCUAGUCCCUACGAUCAGCGUCGUUUACCAACUUCAUCUGAAAUGAACGGC ---(.((((((.......(((((....)))))......(((..((((.(((.(((((.....(((........))).....))))))))))))...)))..........)))))).)... ( -29.50) >consensus ___AUACAUAUAGUCUACGCACCCAAUGGUGCUAUUCCUGGCCACGAACGCAUGAUCCAAUCGACACCAGCUAGUCCCUACGAUCAGCGUCGUUUACCAACUUCAUCUGAAAUGAACGGC ..........(((.....(((((....))))).....)))((((((.((((.(((((.....(((........))).....))))))))))))........(((((.....))))).))) (-27.15 = -26.52 + -0.64)


| Location | 16,184,980 – 16,185,096 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -30.85 |
| Energy contribution | -30.13 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16184980 116 + 22407834 GCCACGAACGCAUGAUCCAAUCGACACCAGCUAGUCCCUACGAUCAGCGUCGUUUACCAACUUCAUCUGAUAUGAACGGUCUAGCCGAAUCAUUGAUUGGAGGGGUAAGUACUUAC---- ...((((.(((.(((((.....(((........))).....))))))))))))(((((..(((((((....((((.(((.....)))..)))).)).))))).)))))........---- ( -30.70) >DroGri_CAF1 113209 109 + 1 GCCACGAACGCAUGAUCCAAUCGACACCAGCUAGUCCUUACGAUCAGCGUCGUUUACCAACUUCAUCUGAAAUGAAUGGCCUAGCCGAAUCAUUGAUUGGAGGGGUGAG----------- ...((((.(((.(((((.....(((........))).....))))))))))))(((((..(((((.....(((((.((((...))))..)))))...))))).))))).----------- ( -31.10) >DroEre_CAF1 105450 120 + 1 GCCACGAACGCAUGAUCCAAUCGACACCAGCUAGUCCCUACGAUCAGCGUCGUUUACCAACUUCAUCUGAUAUGAACGGCCUAGCCGAAUCAUUGAUUGGAGGGGUAAGUAAGAAAAAAG ...((((.(((.(((((.....(((........))).....))))))))))))((((..(((((.((..((((((.((((...))))..))))..))..)))))))..))))........ ( -32.00) >DroWil_CAF1 146876 118 + 1 GCCACGAGCGCAUGAUCCAAUCGACACCAGCUAGUCCUUACGAUCAGCGUCGUUUACCAACUUCAUCUGAUAUGAACGGUCUAGCCGAAUCAUUGAUUGGAGGGGUAAGAAUGUCUAA-- ...((((.(((.(((((.....(((........))).....))))))))))))(((((..(((((((....((((.(((.....)))..)))).)).))))).)))))..........-- ( -30.70) >DroYak_CAF1 110285 109 + 1 GCCACGAGCGCAUGAUCCAAUCGACACCAGCUAGUCCCUACGAUCAGCGUCGUUUACCAACUUCAUCUGAUAUGAACGGCCUAGCCGAAUCAUUGAUUGGAGGGGUAAU----------- ...((((.(((.(((((.....(((........))).....))))))))))))(((((..(((((((....((((.((((...))))..)))).)).))))).))))).----------- ( -31.60) >DroAna_CAF1 98932 115 + 1 GCCACGAACGCAUGAUCCAAUCGACACCAGCUAGUCCUUACGAUCAGCGUCGUUUACCAACUUCAUAUGAAAUGAACGGCCUAGCCGAGUCAUUGAUUGGAGGGGUAAGUGU-AAU---- ...((((.(((.(((((.....(((........))).....))))))))))))(((((..(((((.....(((((.((((...))))..)))))...))))).)))))....-...---- ( -33.50) >consensus GCCACGAACGCAUGAUCCAAUCGACACCAGCUAGUCCCUACGAUCAGCGUCGUUUACCAACUUCAUCUGAUAUGAACGGCCUAGCCGAAUCAUUGAUUGGAGGGGUAAGUA___A_____ ...(((.((((.(((((.....(((........))).....))))))))))))(((((..(((((......((((.((((...))))..))))....))))).)))))............ (-30.85 = -30.13 + -0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:23 2006