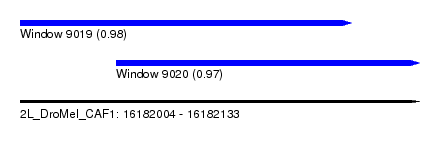
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,182,004 – 16,182,133 |
| Length | 129 |
| Max. P | 0.983192 |

| Location | 16,182,004 – 16,182,111 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.39 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.72 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
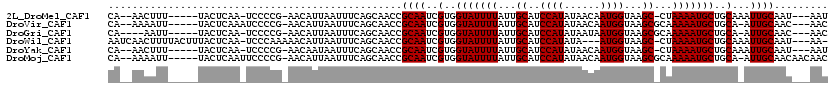
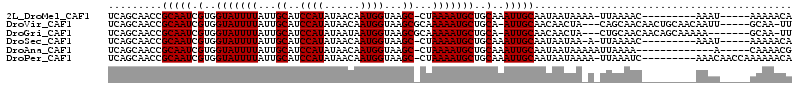
>2L_DroMel_CAF1 16182004 107 + 22407834 CA--AACUUU-----UACUCAA-UCCCCG-AACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAU---AAU ..--......-----.......-......-.....((((((((((.(((((....)))))((((((..((..((((......))))...))-.)))))))))).))))))....---... ( -20.00) >DroVir_CAF1 147491 108 + 1 CA--AAAAUU-----UACUCAAAUCCCCG-AACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGCGCAAAAAUGCUGCA-AUUGCAAC---AAC ..--......-----..............-...................(((((.(..(((((((..(((..((((......))))...)))..)))))))..).-)))))...---... ( -21.80) >DroGri_CAF1 110031 105 + 1 CA----AAUU-----UACUCAA-UCCCCG-AACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAAUAAUGGUAAGCGCAAAAAUGCUGCA-AUUGCAAC---AAC ..----....-----.......-......-...................(((((.(..(((((((..(((..((((......))))...)))..)))))))..).-)))))...---... ( -21.80) >DroWil_CAF1 142630 111 + 1 AAUCAACUUUUACUUUACUCAA-UCCCAAAAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUA---AUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAU---AA- ......................-............((((((((((.(((((....)))))((((((..((..((((...---))))...))-.)))))))))).))))))....---..- ( -18.70) >DroYak_CAF1 107324 107 + 1 CA--AACUUU-----UACUCAA-UCCCCG-AACAAUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAU---AAU ..--......-----.......-......-.....((((((((((.(((((....)))))((((((..((..((((......))))...))-.)))))))))).))))))....---... ( -20.10) >DroMoj_CAF1 166190 111 + 1 CA--AAAAUU-----UACUCAAUUCCCCG-AACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGCGCAAAAAUGCUGCA-AUUGCAACAACAAC ..--......-----..............-...................(((((.(..(((((((..(((..((((......))))...)))..)))))))..).-)))))......... ( -21.80) >consensus CA__AAAAUU_____UACUCAA_UCCCCG_AACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC_CAAAAAUGCUGCA_AUUGCAAC___AAC .................................................((((..(..(((((((...((..((((......))))...))...)))))))..)...))))......... (-17.72 = -17.72 + -0.00)
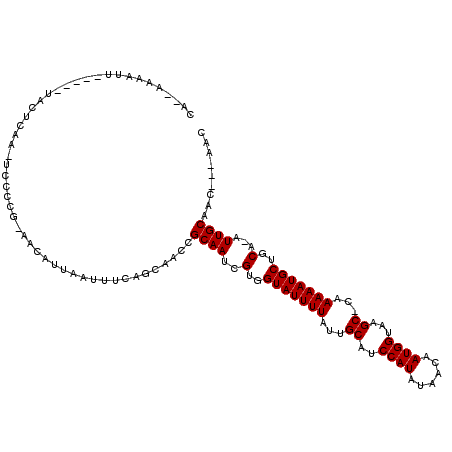
| Location | 16,182,035 – 16,182,133 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.55 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16182035 98 + 22407834 UCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAUAAUAAAA-UUAAAAC---------AAAU-----AAAAACA .........(((((.(..((((((((..((..((((......))))...))-.))))))))..)..)))))..........-.......---------....-----....... ( -19.80) >DroVir_CAF1 147523 104 + 1 UCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGCGCAAAAAUGCUGCA-AUUGCAACAACUA---CAGCAACAACUGCAACAAUU-----GCAA-UU ...((((..(((((.(..(((((((..(((..((((......))))...)))..)))))))..).-)))))........---..(((.....))).....))-----))..-.. ( -28.10) >DroGri_CAF1 110060 102 + 1 UCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAAUAAUGGUAAGCGCAAAAAUGCUGCA-AUUGCAACAACUA---CUGCAACAACAGCAAAAA-------GCAA-UU ...((....(((((.(..(((((((..(((..((((......))))...)))..)))))))..).-)))))........---.(((.......)))....-------))..-.. ( -25.90) >DroSec_CAF1 99037 97 + 1 UCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAUAAUAA-A-UUAAAAC---------AAAU-----AAAAACA .........(((((.(..((((((((..((..((((......))))...))-.))))))))..)..)))))........-.-.......---------....-----....... ( -19.80) >DroAna_CAF1 96339 95 + 1 UCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAUAAUAAAAAUUAAAA-------------A-----CAAAACG .........(((((.(..((((((((..((..((((......))))...))-.))))))))..)..))))).................-------------.-----....... ( -19.80) >DroPer_CAF1 135377 103 + 1 UCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAUAAUAAAA-UUAAAUC---------AAACAACCAAAAAACA .........(((((.(..((((((((..((..((((......))))...))-.))))))))..)..)))))..........-.......---------................ ( -19.80) >consensus UCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC_CUAAAAUGCUGCAAAUUGCAAUAAUAA_A_UUAAAAC_________AAAU_____AAAAACA .........(((((.(..(((((((...((..((((......))))...))...)))))))..)..)))))........................................... (-18.00 = -18.00 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:21 2006