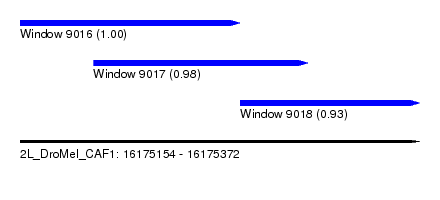
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,175,154 – 16,175,372 |
| Length | 218 |
| Max. P | 0.999991 |

| Location | 16,175,154 – 16,175,274 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -36.15 |
| Consensus MFE | -35.28 |
| Energy contribution | -34.78 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.64 |
| SVM RNA-class probability | 0.999991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
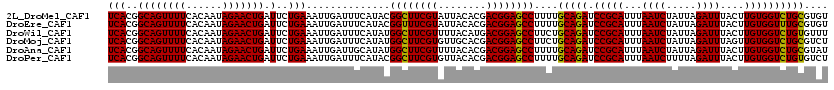
>2L_DroMel_CAF1 16175154 120 + 22407834 UCACGGCAGUUUUCACAAUAGAACUGAUUCUGAAAUUGAUUUCAUACGGCUUCGUAUUACACGACGGAGCCUUUUGCAGAUCCGCAUUUAAUCUAUUAGAUUUACUUGUGGUCUGCGUGU (((..((((((((......))))))).)..)))..........((((((((((((........))))))))....(((((.(((((...((((.....))))....)))))))))))))) ( -38.30) >DroEre_CAF1 95462 120 + 1 UCACGGCAGUUUUCACAAUAGAACUGAUUCUGAAAUUGAUUUCAUACGGUUUCGUAUUACACGACGGAGCCUUUUGCAGAUCCGCAUUUAAUCUAUUAGAUUUACUUGUGGUUUGCGUGU (((..((((((((......))))))).)..)))..........((((((((((((........))))))))....(((((.(((((...((((.....))))....)))))))))))))) ( -33.30) >DroWil_CAF1 134070 120 + 1 UCACGGCAGUUUUCACAAUAGAACUGAUUCUGAAAUUGAUUUCAUAUGGCUUCGUUUUACAUGACGGAGCCUUCUGCAGAUCCGCAUUUAAUCUAUUAGAUUUACUUGUGGUCUGUGUUU (((..((((((((......))))))).)..)))..............(((((((((......)))))))))....(((((.(((((...((((.....))))....)))))))))).... ( -35.70) >DroMoj_CAF1 158259 120 + 1 UCACGGCAGUUUUCACAAUAGAACUGAUUCUGAAAUUGAUUUCAUAUGGCUUCGUGUUGCACGACGGAGCCUUCUGCAGAUCCGCAUUUAAUCUAUUAGAUUUAGUUGUGGUCUGCGUCU (((..((((((((......))))))).)..)))....(((.......((((((((........))))))))....(((((.(((((...((((.....))))....))))))))))))). ( -36.60) >DroAna_CAF1 89367 120 + 1 UCACGGCAGUUUUCACAAUAGAACUGAUUCUGAAAUUGAUUGCAUAUGGCUUCGUUUUACACGACGGAGCCUUUUGCAGAUCCGCAUUUAAUCUAUUAGAUUUACUUGUGGUCUGCGUAU (((..((((((((......))))))).)..))).......(((....(((((((((......)))))))))....(((((.(((((...((((.....))))....))))))))))))). ( -38.40) >DroPer_CAF1 127730 120 + 1 UCACGGCAGUUUUCACAAUAGAACUGAUUCUGAAAUUGAUUUCAUACGGCUUCGUGUUACACGACGGAGCCUUUUGCAGAUCCGCAUUUAAUCUUUUAGAUUUACUUGUGGUCUGUGUCU (((..((((((((......))))))).)..)))....(((.......((((((((........))))))))....(((((.(((((...((((.....))))....))))))))))))). ( -34.60) >consensus UCACGGCAGUUUUCACAAUAGAACUGAUUCUGAAAUUGAUUUCAUACGGCUUCGUAUUACACGACGGAGCCUUUUGCAGAUCCGCAUUUAAUCUAUUAGAUUUACUUGUGGUCUGCGUCU (((..((((((((......))))))).)..)))..............((((((((........))))))))....(((((.(((((...((((.....))))....)))))))))).... (-35.28 = -34.78 + -0.50)
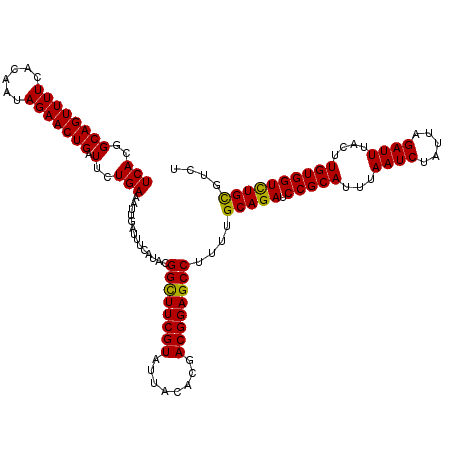
| Location | 16,175,194 – 16,175,311 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -29.42 |
| Energy contribution | -29.28 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
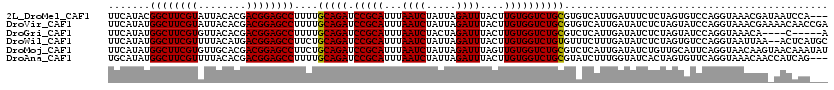
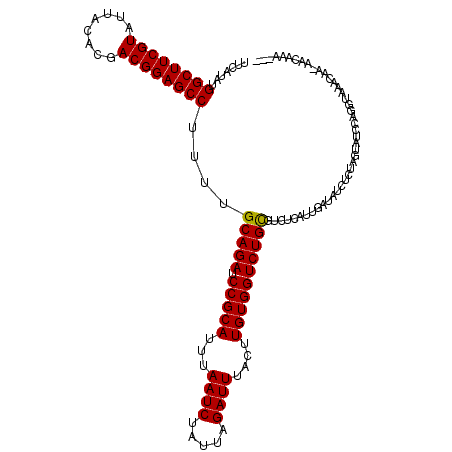
>2L_DroMel_CAF1 16175194 117 + 22407834 UUCAUACGGCUUCGUAUUACACGACGGAGCCUUUUGCAGAUCCGCAUUUAAUCUAUUAGAUUUACUUGUGGUCUGCGUGUCAUUGAUUUCUCUAGUGUCCAGGUAAACGAUAAUCCA--- ...((((((((((((........))))))))....(((((.(((((...((((.....))))....)))))))))))))).((((.(((..((.......))..)))))))......--- ( -32.70) >DroVir_CAF1 140016 120 + 1 UUCAUAUGGCUUCGUAUUACACGACGGAGCCUUUUGCAGAUCCGCAUUUAAUCUAUUAGAUUUACUUGUGGUCUGCGUGUCAUUGAUAUCUCUAGUAUCCAGGUAAACGAAAACAACCGA (((....((((((((........))))))))....(((((.(((((...((((.....))))....))))))))))((..(...(((((.....)))))...)...)))))......... ( -33.10) >DroGri_CAF1 102316 111 + 1 UUCAUAUGGCUUCGUGUUACACGACGGAGCCUUUUGCAGAUCCGCAUUUAAUCUACUAGAUUUACUUGUGGUCUGCGUCUCAUUGAUAUCUCUAGUAUCCAGGUAAACA----C-----A .......((((((((........))))))))....(((((.(((((...((((.....))))....))))))))))((..(...(((((.....)))))...)...)).----.-----. ( -31.50) >DroWil_CAF1 134110 118 + 1 UUCAUAUGGCUUCGUUUUACAUGACGGAGCCUUCUGCAGAUCCGCAUUUAAUCUAUUAGAUUUACUUGUGGUCUGUGUUUCUUUGAUAUCUCUAGUGUCCAGGUAAUUAA--ACUCAUGC .......(((((((((......)))))))))....(((((.(((((...((((.....))))....))))))))))(((.(((.(((((.....))))).))).)))...--........ ( -32.50) >DroMoj_CAF1 158299 120 + 1 UUCAUAUGGCUUCGUGUUGCACGACGGAGCCUUCUGCAGAUCCGCAUUUAAUCUAUUAGAUUUAGUUGUGGUCUGCGUCUCAUUGAUAUCUGUUGCAUUCAGGUAACAAGUAACAAAUAU .......((((((((........))))))))....(((((.(((((...((((.....))))....))))))))))......(((.((..((((((......))))))..)).))).... ( -38.00) >DroAna_CAF1 89407 117 + 1 UGCAUAUGGCUUCGUUUUACACGACGGAGCCUUUUGCAGAUCCGCAUUUAAUCUAUUAGAUUUACUUGUGGUCUGCGUAUCUUUGGUAUCACUAGUGUUCAGGUAAACAACCAUCAG--- .((((..(((((((((......)))))))))....(((((.(((((...((((.....))))....))))))))))..................))))...(((.....))).....--- ( -34.60) >consensus UUCAUAUGGCUUCGUAUUACACGACGGAGCCUUUUGCAGAUCCGCAUUUAAUCUAUUAGAUUUACUUGUGGUCUGCGUCUCAUUGAUAUCUCUAGUAUCCAGGUAAACAA_AACAAA___ .......((((((((........))))))))....(((((.(((((...((((.....))))....))))))))))............................................ (-29.42 = -29.28 + -0.14)

| Location | 16,175,274 – 16,175,372 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -16.52 |
| Consensus MFE | -10.77 |
| Energy contribution | -11.27 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
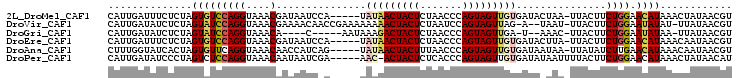
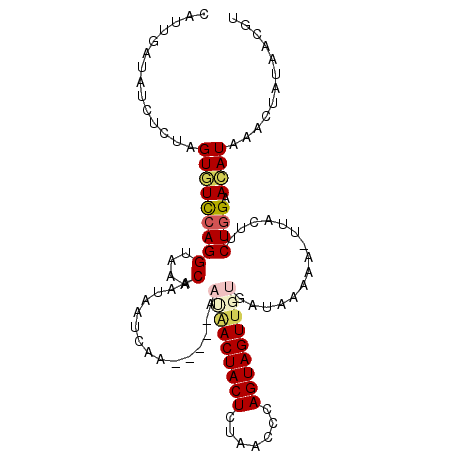
>2L_DroMel_CAF1 16175274 98 + 22407834 CAUUGAUUUCUCUAGUGUCCAGGUAAACGAUAAUCCA-----UAUAACUACUCUAACCCAGUAGUUGUGAUACUAA-UUACUUCUGGAACAUAAACUAUAACGU ..............((((((((((((..(......).-----((((((((((.......)))))))))).......-))))..)))).))))............ ( -17.00) >DroVir_CAF1 140096 99 + 1 CAUUGAUAUCUCUAGUAUCCAGGUAAACGAAAACAACCGAAAAAAAACUACUCUAAUCCAGUAGUUAG-A--UAAU-UUACUUCUGGAAUAUAU-UUAUAACGU .................((((((((((((........))......(((((((.......)))))))..-.--...)-))))..)))))......-......... ( -14.10) >DroGri_CAF1 102396 90 + 1 CAUUGAUAUCUCUAGUAUCCAGGUAAACA----C-----AAUAAAGACUACUCUAACCCAGUAGUUGA-U--AAAC-UUACUUCUGGAAUAUAA-UUAUAACGU .................((((((......----.-----......(((((((.......)))))))..-.--....-.....))))))......-......... ( -13.81) >DroEre_CAF1 95582 98 + 1 CAUUGAUUUCUCUAGUGUCCAGGUAAACGAUAAUCCA-----UAUAACUACUCUAACCCAGUAGUUGUGAUACUUA-UUACUUCUGGAACAUAAACAAUAACGU ..............(((((((((.....(((((....-----((((((((((.......))))))))))....)))-))...))))).))))............ ( -19.10) >DroAna_CAF1 89487 98 + 1 CUUUGGUAUCACUAGUGUUCAGGUAAACAACCAUCAG-----UAUAACUACUUUAACCCAGUAGUUGUGAUAAUAA-UUAUAUCUUGAACAUAAACAAUAACGU ..............(((((((((..............-----((((((((((.......)))))))))).......-......)))))))))............ ( -19.73) >DroPer_CAF1 127850 98 + 1 CAUUGAUAUCCCUAGUCUCCAGGUAAACAAUAAUCGA-----AAC-ACUACUCUCACCCAGUAGUUGUGAUAUAAUUUUACUUCUGGAACAUAAACUAUAACAU ..............((.((((((((((.....((((.-----...-((((((.......))))))..)))).....)))))..))))))).............. ( -15.40) >consensus CAUUGAUAUCUCUAGUGUCCAGGUAAACAAUAAUCAA_____AAUAACUACUCUAACCCAGUAGUUGUGAUAAAAA_UUACUUCUGGAACAUAAACUAUAACGU ..............(((((((((....)...............(((((((((.......)))))))))...............)))).))))............ (-10.77 = -11.27 + 0.50)
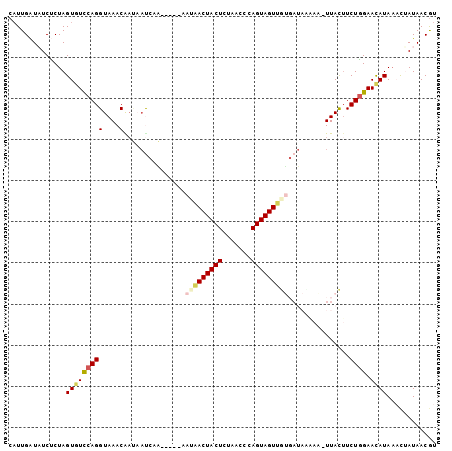
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:19 2006