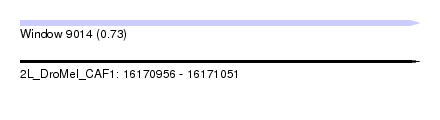
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,170,956 – 16,171,051 |
| Length | 95 |
| Max. P | 0.726897 |

| Location | 16,170,956 – 16,171,051 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -15.88 |
| Consensus MFE | -10.76 |
| Energy contribution | -10.65 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
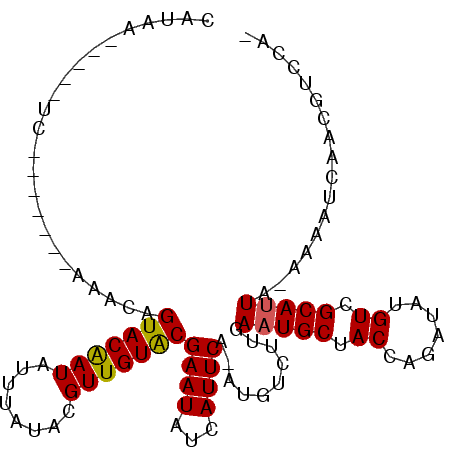
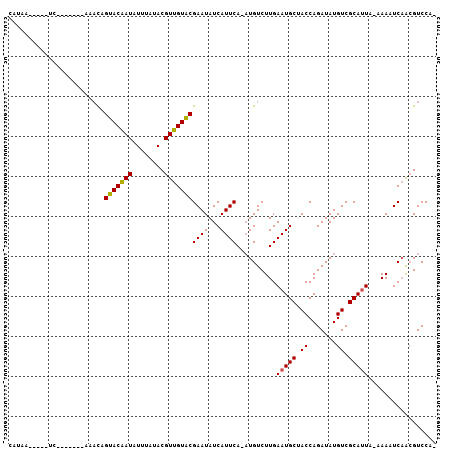
>2L_DroMel_CAF1 16170956 95 + 22407834 CAUAA-----UC-------GAAUCGUACAAUUUUUAUACGUUGUACGAAUAUCAUUCAAAUGUCUUGAAUGCCACCAGAUAUGUCGCAUUUCAAAAUCAACGUCCAC ((((.-----((-------...(((((((((........)))))))))....(((((((.....)))))))......))))))........................ ( -17.00) >DroVir_CAF1 135191 93 + 1 UAUUAUAA-----------AAUCAGUACAAUAUUUAUACGUUGUACGAAUAUCAUUCA-AUGUCUUGAAUGCUACCAGAUAUGUCGCAUUA-AAAUUCAACGUCCU- ........-----------.....(((((((........)))))))((((..((((((-(....)))))))......((....))......-..))))........- ( -13.00) >DroGri_CAF1 97638 104 + 1 AAAACCAACAACAUCUAAACAACAGUACAAUAUUUAUACGUUGUACGAAUAUCAUUCA-AUGUCUUGAAUGCUACCAGAUAUGUCGCACUA-AAAUUCAACGUCCU- ............((((........(((((((........)))))))......((((((-(....))))))).....))))...........-..............- ( -13.40) >DroMoj_CAF1 152890 97 + 1 UAAAAAAACAAC-------AAACAGUACAAUCUUUAUACGUUGUACGAAUAUCAUUCA-AUGUCUUGAAUGCUACCAGAUAUGUCGCAUUA-AAAUUCAACGUCCU- ...........(-------((((((((((((........)))))))((((...)))).-.))).)))(((((.((.......)).))))).-..............- ( -12.80) >DroAna_CAF1 85161 91 + 1 CA--------UC-------AAAUCGUACAAUAUUUAUACGUUGUGCGAAUAUCAUUCA-AUGUCUUGAAUGCCACCAGAUAUGUCGCAUUAAAAAAUCUGAGUCCAA ..--------..-------...(((((((((........)))))))))....((((((-(....)))))))..(((((((...............))))).)).... ( -17.76) >DroPer_CAF1 123536 94 + 1 -GUCA-----UC-------GAAUCGUACGAUGUUUAUACGUUGUACGAAUAUCAUUCAAUUGUCUUGAAUGCUACCAGAUAUGUCGCAUUACAAAAUCAACGUCCAA -((.(-----((-------((.(((((((((((....)))))))))))....(((((((.....)))))))............))).)).))............... ( -21.30) >consensus CAUAA_____UC_______AAACAGUACAAUAUUUAUACGUUGUACGAAUAUCAUUCA_AUGUCUUGAAUGCUACCAGAUAUGUCGCAUUA_AAAAUCAACGUCCA_ ........................(((((((........)))))))((((...))))..........(((((.((.......)).)))))................. (-10.76 = -10.65 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:15 2006