
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,170,099 – 16,170,242 |
| Length | 143 |
| Max. P | 0.907864 |

| Location | 16,170,099 – 16,170,202 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.45 |
| Mean single sequence MFE | -25.85 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.07 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
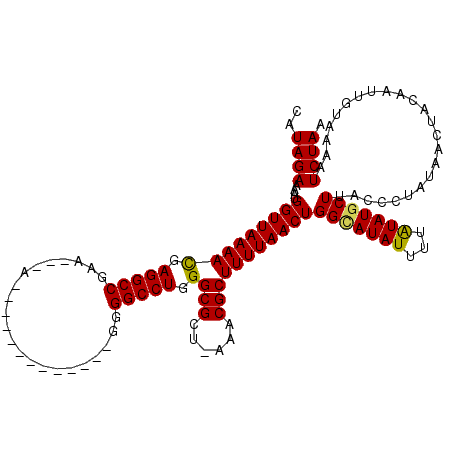
>2L_DroMel_CAF1 16170099 103 - 22407834 CAUAGAAAAUGGUUAAAACGAGGCCGCA---A-------------UGGGCCUGGGCGCU-AAGCGCUUUUAACUGGUAUAUUUAAUAUGCUUACCCUAUAACUACAAUUGUAAAAUCUAA ..((((....(((((((((.(((((...---.-------------..))))).)((((.-..))))))))))))(((((((...)))))))........................)))). ( -26.00) >DroVir_CAF1 134143 105 - 1 CAUAGAAAAUGGUUAAAACGAGGCCGAU---UAGA-----------UGGCCUGGGCGCU-UAACGCUUUUAACUGGCAUAUUUUAUAUGCUUACCCUAUAACUACAAUUGUAAAAUCUAA ..((((....(((((((((.((((((..---....-----------)))))).)(((..-...)))))))))))(((((((...)))))))........................)))). ( -25.40) >DroGri_CAF1 96522 104 - 1 CAUAGAAAAUGGUUAAAAUGAGGCCGGA---A------------UGGGGCCUGGGCGCU-UAACGCUUUUAACUGGCAUAUUUUAUAUGCUUACCCUAUAACUACAAUUGUAAAAUCUAA ..((((....(((((((((.(((((...---.------------...))))).)(((..-...)))))))))))(((((((...)))))))........................)))). ( -23.60) >DroWil_CAF1 127893 107 - 1 CAUAGAAAAUGGUUAAAACGAGGCCUUACAAA-------------GAGGCCUGGGCGCUUAAUCGCUUUUAACUGGCAUAUUUUGUAUGCUUACCCUAUAACUACAAUUGUAAAAUCUAA ..((((....(((((((((.(((((((.....-------------))))))).)(((......)))))))))))(((((((...)))))))........................)))). ( -27.80) >DroMoj_CAF1 151649 116 - 1 CAUAGAAAAUGGUUAAAACUAGGCCGAA---UAUAUAUAUAUUUUUUGGCCUGGGCGCU-CAACGCUUUUAACUGGCAUAUUUUAUAUGCUUACCCUAUAACUACAAUUGUAAAAUCUAA ..((((....((((((((((((((((((---.(((....)))..))))))))))(((..-...)))))))))))(((((((...)))))))........................)))). ( -29.70) >DroPer_CAF1 122506 103 - 1 CAUAGAAAAUGGUUAAAACGAGGCCAGA---G-------------GGGGCCUAGGCGCU-CUGCGCUUUUAACUGGAAUAUUUUGUAUGCUUACCCUAUAACUACAAUUGUAAAAUCUAA ((((.((((((((((((...(((((...---.-------------..))))).(((((.-..)))))))))))).....))))).)))).............(((....)))........ ( -22.60) >consensus CAUAGAAAAUGGUUAAAACGAGGCCGAA___A_____________GGGGCCUGGGCGCU_AAACGCUUUUAACUGGCAUAUUUUAUAUGCUUACCCUAUAACUACAAUUGUAAAAUCUAA ..((((....(((((((((.(((((......................))))).)(((......)))))))))))(((((((...)))))))........................)))). (-22.37 = -22.07 + -0.30)
| Location | 16,170,139 – 16,170,242 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -17.00 |
| Consensus MFE | -15.09 |
| Energy contribution | -15.09 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
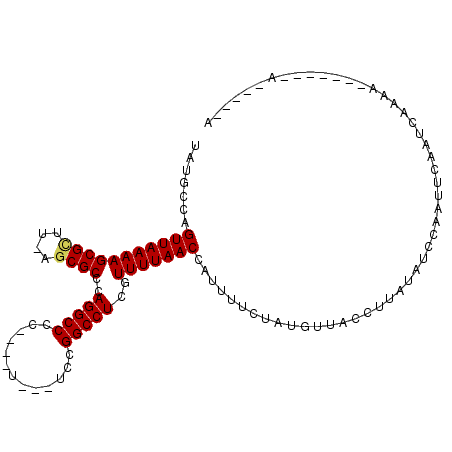
>2L_DroMel_CAF1 16170139 103 + 22407834 UAUACCAGUUAAAAGCGCUU-AGCGCCCAGGCCCA----U---UGCGGCCUCGUUUUAACCAUUUUCUAUGUUACCUUAUAUGCAAUUCAAUCGAAAUUGAUCAAAUCAGA .......(((((((((((..-.))))..(((((..----.---...)))))..)))))))...........................(((((....))))).......... ( -17.40) >DroVir_CAF1 134183 91 + 1 UAUGCCAGUUAAAAGCGUUA-AGCGCCCAGGCCA--UCUA---AUCGGCCUCGUUUUAACCAUUUUCUAUGUUACCUUAUAUCCAAUUCAAUCGAAA-------------- .......((((((((((...-..)))..(((((.--....---...)))))..))))))).....................................-------------- ( -15.30) >DroGri_CAF1 96562 90 + 1 UAUGCCAGUUAAAAGCGUUA-AGCGCCCAGGCCCCA---U---UCCGGCCUCAUUUUAACCAUUUUCUAUGUUACCUUAUAUCCAAUUCAAUCAAAA-------------- .......((((((((((...-..)))..(((((...---.---...)))))..))))))).....................................-------------- ( -14.50) >DroWil_CAF1 127933 95 + 1 UAUGCCAGUUAAAAGCGAUUAAGCGCCCAGGCCUC----UUUGUAAGGCCUCGUUUUAACCAUUUUCUAUGUUACCUUAUAUCCAAUUUAAUCGGCA-------A-----A ..((((.((((((((((......)))..((((((.----......))))))..)))))))(((.....)))......................))))-------.-----. ( -21.20) >DroAna_CAF1 84319 104 + 1 UAUUCCAGUUAAAAGCGCUU-AGCGCCCAGGCCCAU---C---UCCGGCCUCGUUUUAACCAUUUUCUAUGUUACCUUAUAUCCAAUUCAAUCAAAAUUUGCCCAAUCGGA ...(((.(((((((((((..-.))))..(((((...---.---...)))))..)))))))(((.....))).....................................))) ( -17.60) >DroPer_CAF1 122546 103 + 1 UAUUCCAGUUAAAAGCGCAG-AGCGCCUAGGCCCC----C---UCUGGCCUCGUUUUAACCAUUUUCUAUGUUACCUUAUAUCCAAUUUAAUCAAAAUCGAGCCAAUUAGA .......(((((((((((..-.))))..(((((..----.---...)))))..))))))).....................((.((((...((......))...)))).)) ( -16.00) >consensus UAUGCCAGUUAAAAGCGCUU_AGCGCCCAGGCCCC____U___UCCGGCCUCGUUUUAACCAUUUUCUAUGUUACCUUAUAUCCAAUUCAAUCAAAA_______A_____A .......(((((((((((....))))..(((((.............)))))..)))))))................................................... (-15.09 = -15.09 + -0.00)


| Location | 16,170,139 – 16,170,242 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.34 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16170139 103 - 22407834 UCUGAUUUGAUCAAUUUCGAUUGAAUUGCAUAUAAGGUAACAUAGAAAAUGGUUAAAACGAGGCCGCA---A----UGGGCCUGGGCGCU-AAGCGCUUUUAACUGGUAUA ...((((..(((......)))..)))).......................(((((((((.(((((...---.----..))))).)((((.-..))))))))))))...... ( -27.10) >DroVir_CAF1 134183 91 - 1 --------------UUUCGAUUGAAUUGGAUAUAAGGUAACAUAGAAAAUGGUUAAAACGAGGCCGAU---UAGA--UGGCCUGGGCGCU-UAACGCUUUUAACUGGCAUA --------------.((((((...))))))....................(((((((((.((((((..---....--)))))).)(((..-...)))))))))))...... ( -19.00) >DroGri_CAF1 96562 90 - 1 --------------UUUUGAUUGAAUUGGAUAUAAGGUAACAUAGAAAAUGGUUAAAAUGAGGCCGGA---A---UGGGGCCUGGGCGCU-UAACGCUUUUAACUGGCAUA --------------....................................(((((((((.(((((...---.---...))))).)(((..-...)))))))))))...... ( -16.50) >DroWil_CAF1 127933 95 - 1 U-----U-------UGCCGAUUAAAUUGGAUAUAAGGUAACAUAGAAAAUGGUUAAAACGAGGCCUUACAAA----GAGGCCUGGGCGCUUAAUCGCUUUUAACUGGCAUA .-----.-------(((((.(((((...(((.((((((..(((.....))).......(.(((((((.....----))))))).))).)))))))...))))).))))).. ( -26.80) >DroAna_CAF1 84319 104 - 1 UCCGAUUGGGCAAAUUUUGAUUGAAUUGGAUAUAAGGUAACAUAGAAAAUGGUUAAAACGAGGCCGGA---G---AUGGGCCUGGGCGCU-AAGCGCUUUUAACUGGAAUA (((((((.((((.....)).)).)))))))....................(((((((((.(((((...---.---...))))).)((((.-..))))))))))))...... ( -27.30) >DroPer_CAF1 122546 103 - 1 UCUAAUUGGCUCGAUUUUGAUUAAAUUGGAUAUAAGGUAACAUAGAAAAUGGUUAAAACGAGGCCAGA---G----GGGGCCUAGGCGCU-CUGCGCUUUUAACUGGAAUA (((((((...(((....)))...)))))))....................(((((((...(((((...---.----..))))).(((((.-..))))))))))))...... ( -23.50) >consensus U_____U_______UUUCGAUUGAAUUGGAUAUAAGGUAACAUAGAAAAUGGUUAAAACGAGGCCGGA___A____GGGGCCUGGGCGCU_AAGCGCUUUUAACUGGCAUA ..................................................(((((((((.(((((.............))))).)(((......)))))))))))...... (-17.48 = -17.34 + -0.14)

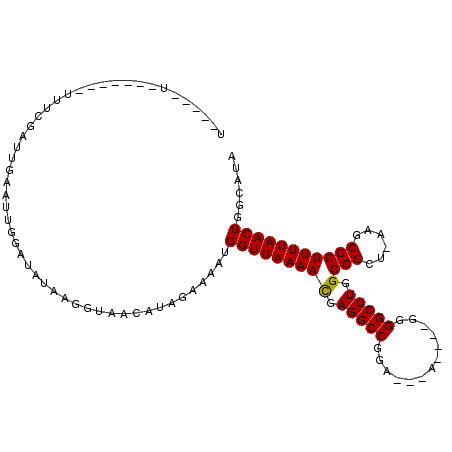
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:14 2006