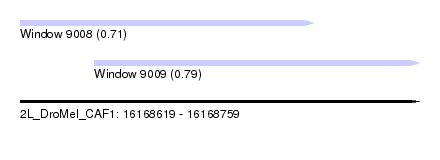
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,168,619 – 16,168,759 |
| Length | 140 |
| Max. P | 0.789455 |

| Location | 16,168,619 – 16,168,722 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.45 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -18.39 |
| Energy contribution | -17.67 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
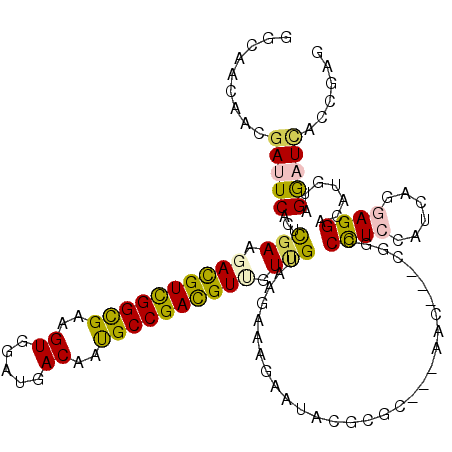
>2L_DroMel_CAF1 16168619 103 + 22407834 UCCGCAUCAUUCAGGAUUGCUGCGGCGGCAACAACGACUCACUCGAAGACGUUGGCGAGGUGGACGACAACGCCGACGUUGUCGUGAGAAAGAACUCAAGG---AA ((((((.((........)).))).((((((((..(((.....))).....(((((((..((.....))..)))))))))))))))(((......)))..))---). ( -35.20) >DroPse_CAF1 131431 97 + 1 UGCGGAUCGUGCGGGCUAG------UGGCAACAAUGAUUCACUCGAGGAUGUCGGCGAAGUGGAUGACAAUGCCGACGUGGUUGUAAGGAAAAAUACGCGC---AA ........(((((......------((....))....(((...(((..(((((((((...((.....)).)))))))))..)))....))).....)))))---.. ( -28.90) >DroEre_CAF1 88960 103 + 1 UCCGCAUCAUUCAGGAUAGCUGCGGUGGCAACAACGACUCUCUCGAAGACGUUGGUGAGGUGGACGACAACGCCGACGUUGUUGUGAGAAAGAACUCGAGG---AA ((((((((......))).))..((.((....)).))..((((.(((.((((((((((..((.....))..)))))))))).))).))))..........))---). ( -33.50) >DroWil_CAF1 125952 103 + 1 UACGAAUUAUUCGCUGUAGAAGCGGUGG---CAAUGAUUCCCUUGAAGACGUUGGCGAUGUGGAUGACAAUGCCGAUGUUGUUGUAAAAAAGAAUACACGCAAUAA ...((((((((((((((....)))))))---..))))))).......((((((((((.(((.....))).))))))))))((((((........)))).))..... ( -28.00) >DroAna_CAF1 82343 100 + 1 UACGGAUCAUCCAAGAAAG---UGGUGGGAACAACGACUCACUCGAGGACGUCGGCGAGGUCGAUGACAACGCCGAUGUUGUGGUGAGAAAGAAUUCGCGC---AA ...((.....)).......---..(((.(((......((((((..(.((((((((((..(((...)))..)))))))))).)))))))......))).)))---.. ( -35.60) >DroPer_CAF1 120461 97 + 1 UGCGGAUCGUGCGGGCUAG------UGGCAACAAUGAUUCACUCGAGGAUGUCGGCGAAGUGGAUGACAAUGCCGACGUGGUUGUAAGGAAAAAUACGCGC---AA ........(((((......------((....))....(((...(((..(((((((((...((.....)).)))))))))..)))....))).....)))))---.. ( -28.90) >consensus UACGGAUCAUUCAGGAUAG___CGGUGGCAACAACGACUCACUCGAAGACGUCGGCGAGGUGGAUGACAACGCCGACGUUGUUGUAAGAAAGAAUACGCGC___AA .........................((....))....((..((((..((((((((((..((.....))..))))))))))....))))..)).............. (-18.39 = -17.67 + -0.72)


| Location | 16,168,645 – 16,168,759 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.02 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -23.69 |
| Energy contribution | -23.12 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16168645 114 + 22407834 GGCAACAACGACUCACUCGAAGACGUUGGCGAGGUGGACGACAACGCCGACGUUGUCGUGAGAAAGAACUCAAGG---AAU---CGUCCCUCGAUCAGAAGGACAUGCAGGAUAACCGAG .(((.....((.((...(((.((((((((((..((.....))..)))))))))).)))((((......))))..)---).)---)((((.((.....)).)))).))).((....))... ( -37.70) >DroPse_CAF1 131451 114 + 1 GGCAACAAUGAUUCACUCGAGGAUGUCGGCGAAGUGGAUGACAAUGCCGACGUGGUUGUAAGGAAAAAUACGCGC---AAC---CGGCCCUCCAUCAGGAGGACAUGUAGGAUCACCGAG (....).........((((..((((((((((...((.....)).)))))))((((((((..(........)..))---)))---)...(((((....))))))).......)))..)))) ( -38.70) >DroGri_CAF1 94713 114 + 1 GG---CAACGAUUCGCUCGAAGAUGUUGGUGAUGUGGACGACAAUGCCGAUGUCAUUGUAAGAAGGAAUACGCGA---AAUAAACGGCCCUCCAUACGGAGGACAUGUAGAAUCACUGAG (.---...)(((((((.((..((((((((((.(((.....))).))))))))))...(((........)))....---......))))(((((....))))).......)))))...... ( -31.50) >DroWil_CAF1 125978 114 + 1 GG---CAAUGAUUCCCUUGAAGACGUUGGCGAUGUGGAUGACAAUGCCGAUGUUGUUGUAAAAAAGAAUACACGCAAUAAU---CGUCCUUCGAUAAGAAGGACAUGUAGAAUCACCGAG ((---...((((((....((.((((((((((.(((.....))).))))))))))((((((........)))).)).....)---)(((((((.....))))))).....))))))))... ( -35.20) >DroAna_CAF1 82366 114 + 1 GGGAACAACGACUCACUCGAGGACGUCGGCGAGGUCGAUGACAACGCCGAUGUUGUGGUGAGAAAGAAUUCGCGC---AAC---CGACCCUCGAUCAGGAGGACAUGUAGGAUUACAGAA ....(((....(((..((((((..(((((((..(((...)))..)))))))((((((.((((......)))))))---)))---....))))))....)))....)))............ ( -36.30) >DroPer_CAF1 120481 114 + 1 GGCAACAAUGAUUCACUCGAGGAUGUCGGCGAAGUGGAUGACAAUGCCGACGUGGUUGUAAGGAAAAAUACGCGC---AAC---CGGCCCUCCAUCAGGAGGACAUGUAGGAUCACCGAG (....).........((((..((((((((((...((.....)).)))))))((((((((..(........)..))---)))---)...(((((....))))))).......)))..)))) ( -38.70) >consensus GGCAACAACGAUUCACUCGAAGACGUCGGCGAAGUGGAUGACAAUGCCGACGUUGUUGUAAGAAAGAAUACGCGC___AAC___CGGCCCUCCAUCAGGAGGACAUGUAGGAUCACCGAG .........(((((...(((.((((((((((..((.....))..)))))))))).)))..............................((((......)))).......)))))...... (-23.69 = -23.12 + -0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:10 2006