| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,762,548 – 1,762,651 |
| Length | 103 |
| Max. P | 0.714114 |

| Location | 1,762,548 – 1,762,651 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.66 |
| Mean single sequence MFE | -20.43 |
| Consensus MFE | -11.68 |
| Energy contribution | -10.85 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
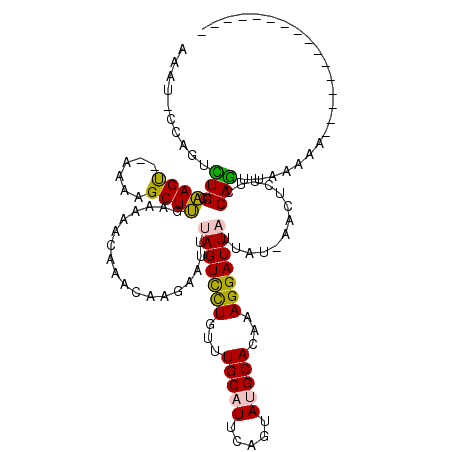
>2L_DroMel_CAF1 1762548 103 + 22407834 AAAUGCCAGUCUGCAAGUUUAAAAGCUUUAAAAACAAUCAAGAAUUUAGUCCUGUUUGCAUUCAGUAUGCACAAAGGAUUUUUAU-AACUCGUCCAGUUAAAUA---------------- ...(((......)))................................((((((...(((((.....)))))...))))))....(-((((.....)))))....---------------- ( -14.30) >DroPse_CAF1 22515 117 + 1 AAAU-CCUGGCUGCGAGC--AAAAGCUCGGAAAACAAACACGAAAUUAGUUUUGUUUGCCUCGGAUAUGCACAAAGGAUUAUUGCCAACGCUUUCAGUUAAAAAGGAAAUCCCACAAAAA .(((-(((...(((((((--....))))((.(((((((.((.......))))))))).))........)))...))))))........................((.....))....... ( -24.70) >DroSec_CAF1 19595 101 + 1 AAAUGCCAGUCUGCAAGU--AAAAGCUUUAAAAACAAACAAGAAUUUAGUCCUGUUUGCAUUCAGUAUGCACAAAGGAUUAUUAU-AACUCGUCCAGUUAAAUC---------------- ...(((......)))...--..........................(((((((...(((((.....)))))...)))))))...(-((((.....)))))....---------------- ( -16.10) >DroEre_CAF1 19470 101 + 1 AAAUCCCAGCUUGUAAGU--AAAAGCUUUAAAAACAAACAGGAAUUUAGUCCUGUUUGCAUUCAGUAUGCAUAAAGGAUUAUUUU-CACUCUUCCAAUCAAACA---------------- .......(((((......--..)))))..............(((..(((((((...(((((.....)))))...)))))))..))-).................---------------- ( -22.00) >DroYak_CAF1 26877 101 + 1 CAAUCCCAGUUUGCAAGU--AAAAGCUUGAAAAACAAAAAAGAAUUAAGUUCUGUUUGCAUUCAGUAUGCACAAAGGAUUAUUUU-CACUCUUUCAGUCAAACA---------------- ........((((((((((--....)))))............(((...((((((...(((((.....)))))...))))))...))-)...........))))).---------------- ( -18.70) >DroPer_CAF1 21532 117 + 1 AAAU-CCUGGCUGCGAGC--AAAAGCUCGGAAAACAAACACGAAAUUAGUUUUGUUUGCCUCGGAUAUGCACAAAGGAUUAUUGCCAACGCUUUCAGUUAACAAGGAAAUCCCACAAAAA ..((-((.(((..(((((--....)))))..(((((((.((.......))))))))))))..)))).........(((((.(((..(((.......)))..)))...)))))........ ( -26.80) >consensus AAAU_CCAGUCUGCAAGU__AAAAGCUUGAAAAACAAACAAGAAUUUAGUCCUGUUUGCAUUCAGUAUGCACAAAGGAUUAUUAU_AACUCUUCCAGUUAAAAA________________ ..........(((.((((......))))..................(((((((...(((((.....)))))...))))))).............)))....................... (-11.68 = -10.85 + -0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:55 2006