
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,147,692 – 16,147,828 |
| Length | 136 |
| Max. P | 0.841844 |

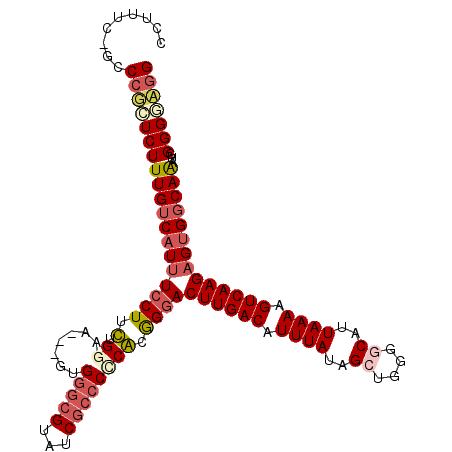
| Location | 16,147,692 – 16,147,802 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -25.55 |
| Energy contribution | -26.53 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
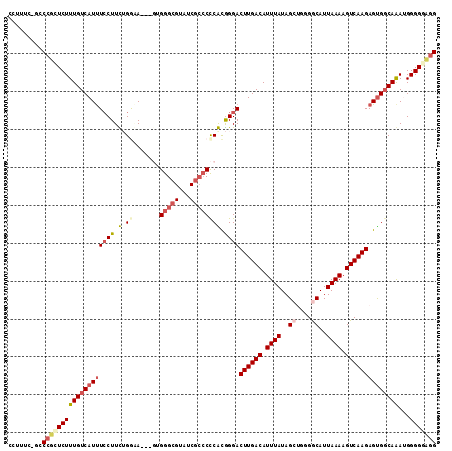
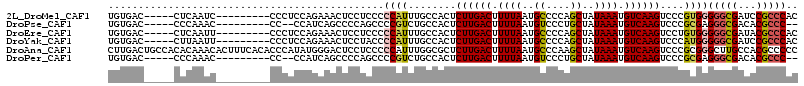
>2L_DroMel_CAF1 16147692 110 + 22407834 CCGUUCAGCCCUCUCUUUGUCAUUUCCUUUUGGAA---GUGGGCGGAUCGCCCCCACGGGACUUGACAUUUAUAGCUGGGGCAUUAAAAGUCAAGAGUGGCAAAUGGGGGAGG ........(((((..((((((((((((....))))---(((((.((....)))))))....((((((.((((..((....))..)))).)))))).)))))))).)))))... ( -41.10) >DroPse_CAF1 109173 102 + 1 CCU------CCGGUCUUUGUCAUUUCCUUCUGGGA-----GGGCGUGUCGCCCUCGCGGGACUUGACAUUUAUAGCAGGGACAUUAAAAGUCAAGAGUGGCAGACGGGGCUGG ...------(((((((((((((((...(((((.((-----(((((...))))))).)))))((((((.((((............)))).))))))))))))))...))))))) ( -42.00) >DroEre_CAF1 72251 109 + 1 CCUUUCCGCCCUCUCUUUGUCAUUUCCUUUGGA-A---GUGGGCGUAUCGCCCCCACAGGACUUGACAUUUAUAGCUGGGGCAUUAAAAGUCAAGAGUGGCAAAUGGGGGAGG (((..(((((((..((.(((((..((((.(((.-.---..(((((...)))))))).))))..))))).....))..))))).......((((....)))).....))..))) ( -44.60) >DroYak_CAF1 79676 110 + 1 CCUUUCCGCCCUCUCUUUGUCAUUUCCUUUGGAAA---GUGGGCGGAUCGCCCCCAUGGGACUUGACAUUUAUAGCUGGGGCAUUAAAAGUCAAGAGUGGCAAAUGGGGUAGG .....(((((((...(((((((((((((.(((...---..(((((...)))))))).))))((((((.((((..((....))..)))).))))))))))))))).))))).)) ( -40.90) >DroAna_CAF1 68348 110 + 1 ---UCGUGCCAGGUCUUUGUCAUCUACUUCUGGUAGACGGGGGCGUGGCAAGCCCGCGGGACUUGACAUUUAUAGCUUGGGCAUUAAAAGUCAAGAGCGCCAAAUGGGGGAGG ---...(((((.((((((((((((.......))).))))))))).)))))..(((((....((((((.((((..((....))..)))).)))))).)).((....)))))... ( -41.40) >DroPer_CAF1 98279 102 + 1 CCU------CCGGUCUUUGUCAUUUCCUUCUGGGA-----GGGCGUGUCGCCCUCGCGGGACUUGACAUUUAUAGCAGGGACAUUAAAAGUCAAGAGUGGCAGACGGGGCUGG ...------(((((((((((((((...(((((.((-----(((((...))))))).)))))((((((.((((............)))).))))))))))))))...))))))) ( -42.00) >consensus CCUUUC_GCCCGCUCUUUGUCAUUUCCUUCUGGAA___GUGGGCGUAUCGCCCCCACGGGACUUGACAUUUAUAGCUGGGGCAUUAAAAGUCAAGAGUGGCAAAUGGGGGAGG .........(((((((((((((((((((.(.((.......(((((...)))))))).))))((((((.((((..((....))..)))).))))))))))))))...))))))) (-25.55 = -26.53 + 0.98)

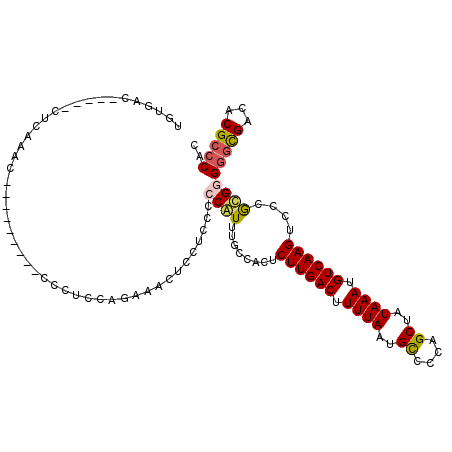
| Location | 16,147,727 – 16,147,828 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -15.43 |
| Energy contribution | -15.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16147727 101 - 22407834 UGUGAC-----CUCAAUC---------CCCUCCAGAAACUCCUCCCCCAUUUGCCACUCUUGACUUUUAAUGCCCCAGCUAUAAAUGUCAAGUCCCGUGGGGGCGAUCCGCCCAC ......-----.......---------..........................((((.((((((.((((..((....))..)))).))))))....))))(((((...))))).. ( -23.20) >DroPse_CAF1 109202 97 - 1 UGUGAC-----CCCAAAC---------CC--CCAUCAGCCCCAGCCCCGUCUGCCACUCUUGACUUUUAAUGUCCCUGCUAUAAAUGUCAAGUCCCGCGAGGGCGACACGCCC-- ......-----.......---------..--......((....(((((((........((((((.((((..((....))..)))).))))))....))).)))).....))..-- ( -18.70) >DroEre_CAF1 72285 101 - 1 UGUGAC-----CUCAAUU---------CCCUCCAGAAACUCCUCCCCCAUUUGCCACUCUUGACUUUUAAUGCCCCAGCUAUAAAUGUCAAGUCCUGUGGGGGCGAUACGCCCAC ......-----.......---------..........................((((.((((((.((((..((....))..)))).))))))....))))(((((...))))).. ( -23.90) >DroYak_CAF1 79711 101 - 1 UGUGAC-----CUUAAUU---------CCCUCCAGAAACUCCUACCCCAUUUGCCACUCUUGACUUUUAAUGCCCCAGCUAUAAAUGUCAAGUCCCAUGGGGGCGAUCCGCCCAC .(((..-----.......---------.................((((((........((((((.((((..((....))..)))).))))))....))))))(((...))).))) ( -20.50) >DroAna_CAF1 68383 115 - 1 CUUGACUGCCACACAAACACUUUCACACCCAUAUGGGACUCCUCCCCCAUUUGGCGCUCUUGACUUUUAAUGCCCAAGCUAUAAAUGUCAAGUCCCGCGGGCUUGCCACGCCCCC .............................((.(((((........))))).))(((..((((((.((((..((....))..)))).))))))...)))((((.......)))).. ( -28.70) >DroPer_CAF1 98308 97 - 1 UGUGAC-----CCCAAAC---------CC--CCAUCAGCCCCAGCCCCGUCUGCCACUCUUGACUUUUAAUGUCCCUGCUAUAAAUGUCAAGUCCCGCGAGGGCGACACGCCC-- ......-----.......---------..--......((....(((((((........((((((.((((..((....))..)))).))))))....))).)))).....))..-- ( -18.70) >consensus UGUGAC_____CUCAAAC_________CCCUCCAGAAACUCCUCCCCCAUUUGCCACUCUUGACUUUUAAUGCCCCAGCUAUAAAUGUCAAGUCCCGCGGGGGCGACACGCCCAC ..............................................((((........((((((.((((..((....))..)))).))))))....))))(((((...))))).. (-15.43 = -15.43 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:01 2006