| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,124,730 – 16,124,822 |
| Length | 92 |
| Max. P | 0.992157 |

| Location | 16,124,730 – 16,124,822 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.38 |
| Mean single sequence MFE | -19.02 |
| Consensus MFE | -10.79 |
| Energy contribution | -11.02 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
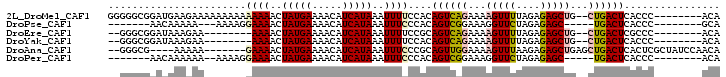
>2L_DroMel_CAF1 16124730 92 + 22407834 GGGGGCGGAUGAAGAAAAAAAAAAAAAACUAUGAAAACAUCAUAAAUUUUCCACAGUCAGAAAAGUUUUAGAGAGCUG--CUGACUCACCC--------ACA (((((..(((...................(((((.....))))).)))..))..((((((...(((((....))))).--))))))..)))--------... ( -18.95) >DroPse_CAF1 86413 79 + 1 -------AACAAAAA---AAAAGGAAAACUAUGAAAACAUCAUAAAUUUCCCACAGUCGGAAAGGUUCUAGAGAGC-----UGACUCACCC--------GCA -------........---....(((((..(((((.....)))))..)))))....((.((...(((((....))))-----)......)).--------)). ( -14.30) >DroEre_CAF1 54689 82 + 1 --GGGCGGAUAAAGAA--------AAAACUAUGAAAACAUCAUAAAUUUUCCGCAGUCAGAAAAGUUUUAGAGAGCUG--CUGACUCGCCC--------ACA --(((((......(((--------((...(((((.....)))))..)))))...((((((...(((((....))))).--)))))))))))--------... ( -26.00) >DroYak_CAF1 61933 82 + 1 --GGGCGGAUAAAGAA--------AAAACUAUGAAAACAUCAUAAAUUUUCCACAGUCAGAAAAGUUUUAGAGAGCUG--CUGACUCACCC--------ACA --(((.(......(((--------((...(((((.....)))))..)))))...((((((...(((((....))))).--))))))).)))--------... ( -20.60) >DroAna_CAF1 51213 89 + 1 --GGGCG----AAAAA-------GAAAACUAUGAAAACAUCAUAAAUUUCCCGCAGUUGGAAAAGUUUAAGAGAGCUGAGCUGACUCACUCGCUAUCCAACA --..(((----.....-------((((..(((((.....)))))..)))).))).((((((..(((......(((.((((....)))))))))).)))))). ( -20.30) >DroPer_CAF1 76442 80 + 1 -------AACAAAAAA--AAAAGGAAAACUAUGAAAACAUCAUAAAUUUCCCACAGUCGGAAAGGUUCUAGAGAGC-----UGACUCACCC--------ACA -------.........--....(((((..(((((.....)))))..)))))....((.((...(((((....))))-----)......)).--------)). ( -14.00) >consensus __GGGCGGAUAAAAAA_______GAAAACUAUGAAAACAUCAUAAAUUUCCCACAGUCAGAAAAGUUUUAGAGAGCUG__CUGACUCACCC________ACA .......................((((..(((((.....)))))..))))....((((((...(((((....)))))...))))))................ (-10.79 = -11.02 + 0.22)

| Location | 16,124,730 – 16,124,822 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 77.38 |
| Mean single sequence MFE | -20.01 |
| Consensus MFE | -11.61 |
| Energy contribution | -10.92 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
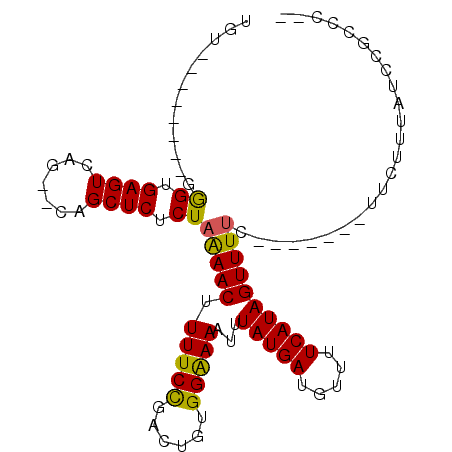
>2L_DroMel_CAF1 16124730 92 - 22407834 UGU--------GGGUGAGUCAG--CAGCUCUCUAAAACUUUUCUGACUGUGGAAAAUUUAUGAUGUUUUCAUAGUUUUUUUUUUUUUUCUUCAUCCGCCCCC .((--------(((((((((((--..................)))))...((((((.((((((.....)))))).)))))).........)))))))).... ( -20.47) >DroPse_CAF1 86413 79 - 1 UGC--------GGGUGAGUCA-----GCUCUCUAGAACCUUUCCGACUGUGGGAAAUUUAUGAUGUUUUCAUAGUUUUCCUUUU---UUUUUGUU------- ..(--------(((.(((...-----.)))...........)))).....((((((.((((((.....)))))).))))))...---........------- ( -15.30) >DroEre_CAF1 54689 82 - 1 UGU--------GGGCGAGUCAG--CAGCUCUCUAAAACUUUUCUGACUGCGGAAAAUUUAUGAUGUUUUCAUAGUUUU--------UUCUUUAUCCGCCC-- ...--------(((((.....(--(((.((..............))))))((((((.((((((.....)))))).)))--------)))......)))))-- ( -22.94) >DroYak_CAF1 61933 82 - 1 UGU--------GGGUGAGUCAG--CAGCUCUCUAAAACUUUUCUGACUGUGGAAAAUUUAUGAUGUUUUCAUAGUUUU--------UUCUUUAUCCGCCC-- .((--------(((((((((((--..................)))))...((((((.((((((.....)))))).)))--------))).))))))))..-- ( -20.67) >DroAna_CAF1 51213 89 - 1 UGUUGGAUAGCGAGUGAGUCAGCUCAGCUCUCUUAAACUUUUCCAACUGCGGGAAAUUUAUGAUGUUUUCAUAGUUUUC-------UUUUU----CGCCC-- .((((((.((.(((((((....)))).))).))........)))))).(((..(((.((((((.....)))))).....-------)))..----)))..-- ( -25.10) >DroPer_CAF1 76442 80 - 1 UGU--------GGGUGAGUCA-----GCUCUCUAGAACCUUUCCGACUGUGGGAAAUUUAUGAUGUUUUCAUAGUUUUCCUUUU--UUUUUUGUU------- .((--------.((.(((...-----.((....))...))).)).))...((((((.((((((.....)))))).))))))...--.........------- ( -15.60) >consensus UGU________GGGUGAGUCAG__CAGCUCUCUAAAACUUUUCCGACUGUGGAAAAUUUAUGAUGUUUUCAUAGUUUUC_______UUCUUUAUCCGCCC__ ............((.((((.......)))).))(((((.(((((......)))))...(((((.....))))))))))........................ (-11.61 = -10.92 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:57 2006