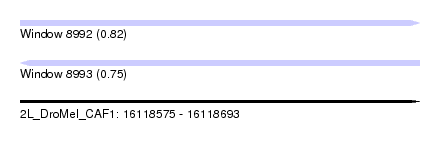
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,118,575 – 16,118,693 |
| Length | 118 |
| Max. P | 0.816338 |

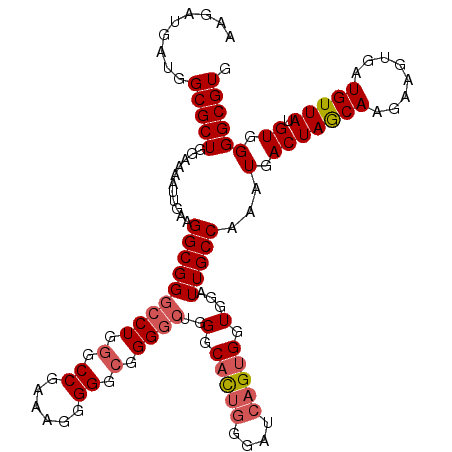
| Location | 16,118,575 – 16,118,693 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.79 |
| Mean single sequence MFE | -36.76 |
| Consensus MFE | -27.92 |
| Energy contribution | -28.80 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16118575 118 + 22407834 AAGAUGAUGGCGCUGGAAAAUUGAAGGCGGGCCUGGGCCGAAAGGGGGCGGGGCUGGGCACCAGGAUCAGUGGUGGAUUGCCAAAUGACUAGCAAGAAGUGAUGUUAUGUGGGGCGUG .........(((((....((((.....(.(((((.(.((......)).).))))).).(((((.......))))))))).(((.(((((..((.....))...))))).)))))))). ( -36.30) >DroSec_CAF1 47715 118 + 1 AAGAUGAUGGCGCUGGAAAAUUGAAGGCGGGCCUGGGCCGAAAGGGGGCGGGGCUGGGCACUGGGAUCAGUGGUGGAUUGCCAAAUGACUAGCAAGAAGUGAUGUUAUGUGGGGCGUG .........(((((....((((.(...(.(((((.(.((......)).).))))).).(((((....))))).).)))).(((.(((((..((.....))...))))).)))))))). ( -36.70) >DroSim_CAF1 48857 118 + 1 AAGAUGAUGGCGCUGGAAAAUUGAAGGCGGGCCUGGGCCGAAAGGGGGCGGGGCUGGGCACUGGGAUCAGUGGUGGAUUGCCAAAUGACUAACAAGAAAUGAUGUUAUGUGGGGCGUG .........(((((...........(((((((((.(.((......)).).))))).(.(((((....))))).)....))))...(.(((((((........))))).)).)))))). ( -35.40) >DroEre_CAF1 49180 117 + 1 AAGAUGAUGGCGCUGGAAAAUUGAAGGCGGAACUGAGCCGGAAU-GGGCGGGCCAGGGCACUGGGAUCAGUGGUGGAUUGCCAAAUGACUAGCAAAAAGUGAUGUUAUGUGGGGCGUG .........(((((.............((..((((((((.....-.)))...((((....))))..)))))..)).....(((.(((((..((.....))...))))).)))))))). ( -32.30) >DroYak_CAF1 51933 116 + 1 AAGAUGAUGGCGCUAGAAAAUUGGAGGCGGGCCUCGGCCGA-AA-GGGCGGGGCAGGGCCUUGGGAUCAGUGGUGGAUUGCCAAAUGACUAGCAAAAAGUGAUGUUAUGUGGGGCGUG .........(((((......((.(((((..((((((.((..-..-)).))))))...))))).))...............(((.(((((..((.....))...))))).)))))))). ( -43.10) >consensus AAGAUGAUGGCGCUGGAAAAUUGAAGGCGGGCCUGGGCCGAAAGGGGGCGGGGCUGGGCACUGGGAUCAGUGGUGGAUUGCCAAAUGACUAGCAAGAAGUGAUGUUAUGUGGGGCGUG .........(((((...........(((((((((.(.((......)).).))))..(.(((((....))))).)...)))))...(.(((((((........))))).)).)))))). (-27.92 = -28.80 + 0.88)


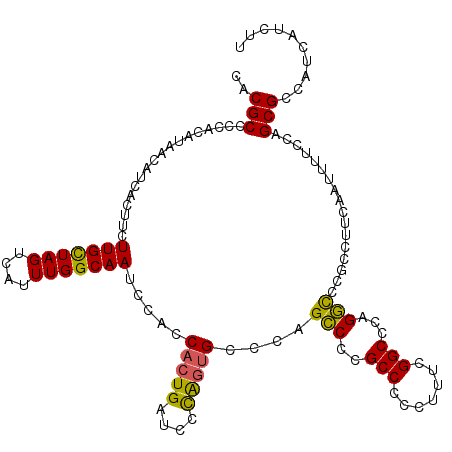
| Location | 16,118,575 – 16,118,693 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 92.79 |
| Mean single sequence MFE | -22.06 |
| Consensus MFE | -17.82 |
| Energy contribution | -17.26 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16118575 118 - 22407834 CACGCCCCACAUAACAUCACUUCUUGCUAGUCAUUUGGCAAUCCACCACUGAUCCUGGUGCCCAGCCCCGCCCCCUUUCGGCCCAGGCCCGCCUUCAAUUUUCCAGCGCCAUCAUCUU .......................(((((((....))))))).........(((..((((((........(((.......)))..(((....)))...........))))))..))).. ( -22.00) >DroSec_CAF1 47715 118 - 1 CACGCCCCACAUAACAUCACUUCUUGCUAGUCAUUUGGCAAUCCACCACUGAUCCCAGUGCCCAGCCCCGCCCCCUUUCGGCCCAGGCCCGCCUUCAAUUUUCCAGCGCCAUCAUCUU ..(((..................(((((((....))))))).....(((((....)))))....(((..(((.......)))...))).................))).......... ( -20.70) >DroSim_CAF1 48857 118 - 1 CACGCCCCACAUAACAUCAUUUCUUGUUAGUCAUUUGGCAAUCCACCACUGAUCCCAGUGCCCAGCCCCGCCCCCUUUCGGCCCAGGCCCGCCUUCAAUUUUCCAGCGCCAUCAUCUU ..(((......(((((........))))).......(((.......(((((....)))))....(((..(((.......)))...)))..)))............))).......... ( -23.20) >DroEre_CAF1 49180 117 - 1 CACGCCCCACAUAACAUCACUUUUUGCUAGUCAUUUGGCAAUCCACCACUGAUCCCAGUGCCCUGGCCCGCCC-AUUCCGGCUCAGUUCCGCCUUCAAUUUUCCAGCGCCAUCAUCUU ..(((..................(((((((....)))))))......(((((..((((....))))...(((.-.....))))))))..................))).......... ( -20.60) >DroYak_CAF1 51933 116 - 1 CACGCCCCACAUAACAUCACUUUUUGCUAGUCAUUUGGCAAUCCACCACUGAUCCCAAGGCCCUGCCCCGCCC-UU-UCGGCCGAGGCCCGCCUCCAAUUUUCUAGCGCCAUCAUCUU .........................(((((....((((...........((....))((((...((((.(((.-..-..))).).)))..))))))))....)))))........... ( -23.80) >consensus CACGCCCCACAUAACAUCACUUCUUGCUAGUCAUUUGGCAAUCCACCACUGAUCCCAGUGCCCAGCCCCGCCCCCUUUCGGCCCAGGCCCGCCUUCAAUUUUCCAGCGCCAUCAUCUU ..(((..................(((((((....))))))).....(((((....)))))....(((..(((.......)))...))).................))).......... (-17.82 = -17.26 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:55 2006