
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,110,402 – 16,110,495 |
| Length | 93 |
| Max. P | 0.979888 |

| Location | 16,110,402 – 16,110,495 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 92.42 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -23.38 |
| Energy contribution | -22.82 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
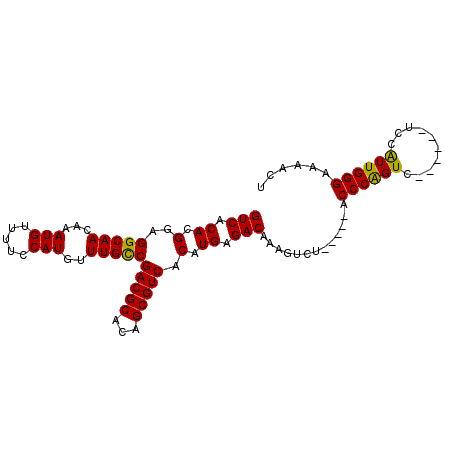
>2L_DroMel_CAF1 16110402 93 + 22407834 GUCACACGGAGGCAACAAAUGUUUUCCAUGUUUGCCGACGCACAGCGUCACAUGAGACAAAGUCUCAUGUUCCCAGUC------UCCAUUGGGAAAACU ..........(((((...(((.....)))..)))))(((((...)))))(((((((((...)))))))))(((((((.------...)))))))..... ( -33.20) >DroSec_CAF1 39534 88 + 1 GUCACACGGAGGCAACAAAUGUUUUCCAUGUUUGCCGACGCACAGCGUCACAUGAGACAAAGUCU-----ACCCGGUC------UCCAUUGGGAAAACU (((.((.(..(((((...(((.....)))..)))))(((((...))))).).)).))).......-----.((((((.------...))))))...... ( -24.40) >DroSim_CAF1 40698 88 + 1 GUCACACGGAGGCAACAAAUGUUUUCCAUGUUUGCCGACGCACAGCGUCACAUGAGACAAAGUCU-----ACCCGGUC------UCCAUUGGGAAAACU (((.((.(..(((((...(((.....)))..)))))(((((...))))).).)).))).......-----.((((((.------...))))))...... ( -24.40) >DroEre_CAF1 40987 94 + 1 GUCACACGGAGGCAACAAAUGUUUGCCAUGUUUGUCGACGCACAGCGUCACAUGAGACAAAGUCU-----ACCCAGUCCGCUGCUCCAUUGGGAAAACU (((.((....((((((....).))))).....(((.(((((...)))))))))).))).......-----.((((((..........))))))...... ( -23.40) >DroYak_CAF1 41596 88 + 1 GUCACACGGAGGCAACAAAUGUUUUCCAUGUUUGCCGACGCACCGCGUCACAUGAGACAAAGUCU-----ACCCAGUC------UCCGUUGGGAAAACU .(((.((((((((.............(((((.....(((((...))))))))))((((...))))-----.....)))------))))))))....... ( -25.60) >consensus GUCACACGGAGGCAACAAAUGUUUUCCAUGUUUGCCGACGCACAGCGUCACAUGAGACAAAGUCU_____ACCCAGUC______UCCAUUGGGAAAACU (((.((.(..(((((...(((.....)))..)))))(((((...))))).).)).))).............((((((..........))))))...... (-23.38 = -22.82 + -0.56)

| Location | 16,110,402 – 16,110,495 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 92.42 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -22.35 |
| Energy contribution | -22.55 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
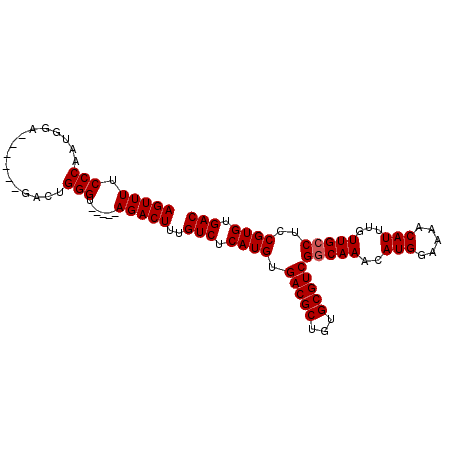
>2L_DroMel_CAF1 16110402 93 - 22407834 AGUUUUCCCAAUGGA------GACUGGGAACAUGAGACUUUGUCUCAUGUGACGCUGUGCGUCGGCAAACAUGGAAAACAUUUGUUGCCUCCGUGUGAC .....(((((..(..------..))))))(((((((((...)))))))))(((((...))))).....(((((((.(((....)))...)))))))... ( -35.50) >DroSec_CAF1 39534 88 - 1 AGUUUUCCCAAUGGA------GACCGGGU-----AGACUUUGUCUCAUGUGACGCUGUGCGUCGGCAAACAUGGAAAACAUUUGUUGCCUCCGUGUGAC (((((.(((...(..------..).))).-----)))))..(((.((((.(((((...)))))(((((..(((.....)))...)))))..)))).))) ( -29.50) >DroSim_CAF1 40698 88 - 1 AGUUUUCCCAAUGGA------GACCGGGU-----AGACUUUGUCUCAUGUGACGCUGUGCGUCGGCAAACAUGGAAAACAUUUGUUGCCUCCGUGUGAC (((((.(((...(..------..).))).-----)))))..(((.((((.(((((...)))))(((((..(((.....)))...)))))..)))).))) ( -29.50) >DroEre_CAF1 40987 94 - 1 AGUUUUCCCAAUGGAGCAGCGGACUGGGU-----AGACUUUGUCUCAUGUGACGCUGUGCGUCGACAAACAUGGCAAACAUUUGUUGCCUCCGUGUGAC .....((...((((((((((((((..((.-----...))..))))((((((((((...))))).....)))))..........)))).))))))..)). ( -27.10) >DroYak_CAF1 41596 88 - 1 AGUUUUCCCAACGGA------GACUGGGU-----AGACUUUGUCUCAUGUGACGCGGUGCGUCGGCAAACAUGGAAAACAUUUGUUGCCUCCGUGUGAC (((((.((((..(..------..))))).-----)))))..(((.((((.(((((...)))))(((((..(((.....)))...)))))..)))).))) ( -31.00) >consensus AGUUUUCCCAAUGGA______GACUGGGU_____AGACUUUGUCUCAUGUGACGCUGUGCGUCGGCAAACAUGGAAAACAUUUGUUGCCUCCGUGUGAC (((((.(((................)))......)))))..(((.((((.(((((...)))))(((((..(((.....)))...)))))..)))).))) (-22.35 = -22.55 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:52 2006