| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,761,012 – 1,761,127 |
| Length | 115 |
| Max. P | 0.543869 |

| Location | 1,761,012 – 1,761,127 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.90 |
| Mean single sequence MFE | -29.27 |
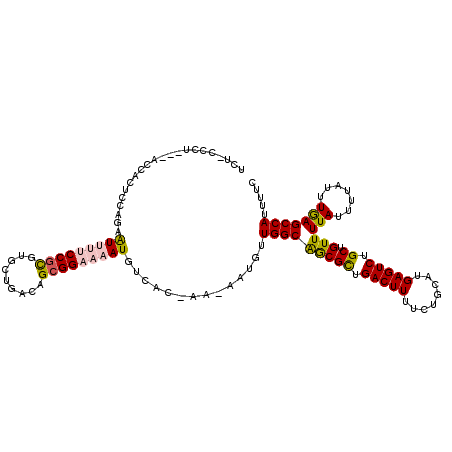
| Consensus MFE | -20.32 |
| Energy contribution | -20.27 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
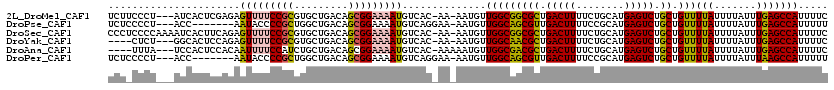
>2L_DroMel_CAF1 1761012 115 + 22407834 UCUUCCCU---AUCACUCGAGAGUUUUCCGCGUGCUGACAGCGGAAAAUGUCAC-AA-AAUGUUGGCGGCGCUGACUUUUCUGCAUGAGUCUGCUGUUUUAUUUUAUUUGAGCCAUUUUC ........---....((((((.(((((((((((....)).))))))))).....-((-((((..(((((((..(((((........)))))))))))).)))))).))))))........ ( -30.50) >DroPse_CAF1 20810 109 + 1 UCUCCCCU---ACC-------AAUACCCCGCUGGCUGACAGCGGAAAAUGUCAGGAA-AAUGUUGGCAGCGUUGACUUUUCCGCAUGAGUCUGCUGUUUUAUUUUAUUUGAGCCAUUUUU ........---...-------..........((((((((((((((..((((..((((-((.((..((...))..))))))))))))...)))))))))............)))))..... ( -31.50) >DroSec_CAF1 18044 118 + 1 CCCUCCCCAAAAUCACUUCAGAGUUUUCCGCGUGCUGACAGCGGAAAAUGUCAC-AA-AAUGUUGGCGGCGCUGACUUUUCUGCAUGAGUCUGCUGUUUUAUUUUAUUUGAGCCAUUUUC ................(((((((((((((((((....)).))))))))).....-((-((((..(((((((..(((((........)))))))))))).)))))).))))))........ ( -28.90) >DroYak_CAF1 25360 111 + 1 ----CUCU---GGCACUCCAGAGUUUUCCGCGUGCUGACAGCGGAAAAUGUCAC-AA-AAUGUUGGCAACGCUGACUUUUCUGCAUGAGUCUGCUGUUUUAUUUUAUUUGAGCCAUUUUC ----((((---((....))))))........(.((((((((((((..(((.((.-((-((.((..((...))..)))))).)))))...)))))))))............))))...... ( -31.50) >DroAna_CAF1 17511 112 + 1 ----UUUA---UCCACUCCACAAUUUUCCAUCUGCUGACAGCGGAAAAUGUCAC-AAAAAUGUUGGCGACGCUGACUUUUCUGCAUGAGUCUGCUGUUUUAUUUUAUUUGAGCCAUUUUC ----....---......................((((((((((((..(((.((.-((((..((..((...))..)))))).)))))...)))))))))............)))....... ( -21.70) >DroPer_CAF1 19834 109 + 1 UCUCCCCU---ACC-------AAUACCCCGCUGGCUGACAGCGGAAAAUGUCAGGAA-AAUGUUGGCAGCGUUGACUUUUCCGCAUGAGUCUGCUGUUUUAUUUUAUUUAAGCCAUUUUU ........---...-------..........((((((((((((((..((((..((((-((.((..((...))..))))))))))))...)))))))))............)))))..... ( -31.50) >consensus UCU_CCCU___ACCACUCCAGAAUUUUCCGCGUGCUGACAGCGGAAAAUGUCAC_AA_AAUGUUGGCAGCGCUGACUUUUCUGCAUGAGUCUGCUGUUUUAUUUUAUUUGAGCCAUUUUC ......................(((((((((.........)))))))))..............(((((((((.(((((........))))).)).)))(((.......)))))))..... (-20.32 = -20.27 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:54 2006