
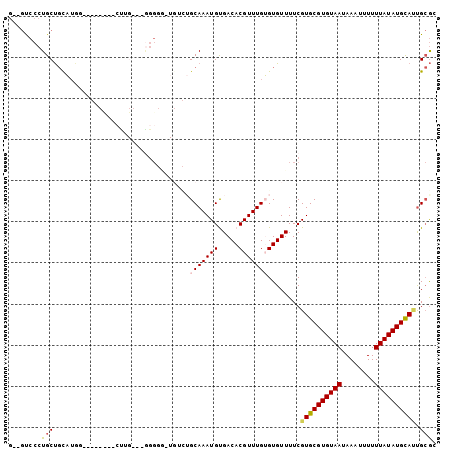
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,100,375 – 16,100,505 |
| Length | 130 |
| Max. P | 0.945741 |

| Location | 16,100,375 – 16,100,476 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -14.73 |
| Energy contribution | -14.87 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16100375 101 + 22407834 G--GUCCCUGCUGCAUAGAAGACCUUCUUGGA-GGGGGCUGUCUGCAAAUGUGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCGG (--(((((..((.((.((((....)))))).)-)))))))..(((((((((.((((((....))))))..)))((((((((........)))))))).)))))) ( -32.70) >DroPse_CAF1 38658 82 + 1 GGGGUCCCUGCUGCCUG---------CU-----GGG--------GCAAAUGUGACACGUUUGGGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGUACUGCGC ...(((((.((.....)---------).-----)))--------))......(((((......)))))...((((((((((........))))))))))..... ( -25.60) >DroYak_CAF1 31782 100 + 1 G--GUCCCUGCUGCAUAGA-GGCCAACUUGGA-AGGGGCUGUCUGCAAAUGCGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCGG (--((((((....(...((-(.....))).).-)))))))..((((((....((((((....))))))....(((((((((........))))))))))))))) ( -30.80) >DroMoj_CAF1 58355 87 + 1 ----CCCCUGCCCCAUUG-------CCUUG------CGAUGGUUGCAAAUGCGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUCGAAC ----....(((.((((((-------(...)------))))))..))).....((((((....))))))(((((((((((((........))))))))).)))). ( -27.20) >DroAna_CAF1 28515 102 + 1 G--GUCCCUGCUGUAAGGAUGGCUGAGCGGAAUGGGGAUUGUCUGCAAAUGUGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAGUGCGG (--((((((.((((..((....))..))))...)))))))..(((((.(((.((((((....))))))..)))((((((((........))))))))..))))) ( -31.30) >DroPer_CAF1 40739 82 + 1 GGGGUCCCUGCUGCCUG---------CU-----GGG--------GCAAAUGUGACACGUUUGGGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGUACUGCGC ...(((((.((.....)---------).-----)))--------))......(((((......)))))...((((((((((........))))))))))..... ( -25.60) >consensus G__GUCCCUGCUGCAUGG________CUUG___GGGGG_UGUCUGCAAAUGUGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCGC ........(((.................................(((((((.....)))))))........((((((((((........)))))))))).))). (-14.73 = -14.87 + 0.14)


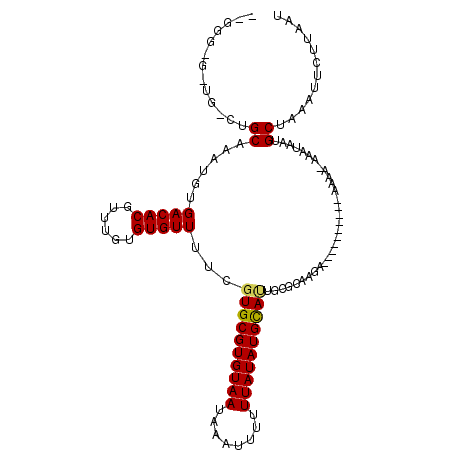
| Location | 16,100,404 – 16,100,505 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.83 |
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -14.99 |
| Energy contribution | -14.77 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16100404 101 + 22407834 A-GGGGGCUGUCUGCAAAUGUGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCGGACG-----------AGCAGAAAUAAUGCUAAAUUCUUAAU (-((((..((((((((((((.((((((....))))))..)))((((((((........)))))))).)))))))))-----------((((.......))))...)))))... ( -29.90) >DroPse_CAF1 38677 95 + 1 --GGG--------GCAAAUGUGACACGUUUGGGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGUACUGCGCAAGA--------CAAAAAAAAAAAAUGCUAAAUUCUUAAU --...--------.((((((.....))))))((((((((.((((((((((........))))))))))(((....).--------)).......))))))))........... ( -20.60) >DroGri_CAF1 30802 89 + 1 -----CGAUGACUGCAAAUGCGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCAAAAAAAA-------------------AGCUAAAUUCUUAAU -----(((.(((((((((((.....)))))))).))).)))(((((((((........)))))))))............-------------------............... ( -18.60) >DroMoj_CAF1 58374 108 + 1 -----CGAUGGUUGCAAAUGCGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUCGAACAAAAAACAGCAACAACAACAAAUAUGGCUAAAUUCUUAAU -----.....(((((......((((((....))))))(((((((((((((........))))))))).)))).........)))))........................... ( -24.20) >DroAna_CAF1 28544 102 + 1 AUGGGGAUUGUCUGCAAAUGUGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAGUGCGGACG-----------AACAAAAAUAAUGCUAAAUUCUUAAU .(((((((((((((((.(((.((((((....))))))..)))((((((((........))))))))..))))))).-----------..((.......))...)))))))).. ( -26.80) >DroPer_CAF1 40758 94 + 1 --GGG--------GCAAAUGUGACACGUUUGGGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGUACUGCGCAAGA--------CAAAAA-AAAAAAUGCUAAAUUCUUAAU --...--------.((((((.....))))))((((((((.((((((((((........))))))))))(((....).--------))....-..))))))))........... ( -20.60) >consensus __GGG_G_UG_CUGCAAAUGUGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCGCAAGA__________AAAA_AAAUAAUGCUAAAUUCUUAAU .............((......(((((......)))))...((((((((((........))))))))))...............................))............ (-14.99 = -14.77 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:48 2006