
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,089,093 – 16,089,235 |
| Length | 142 |
| Max. P | 0.981418 |

| Location | 16,089,093 – 16,089,183 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.34 |
| Mean single sequence MFE | -15.30 |
| Consensus MFE | -9.72 |
| Energy contribution | -9.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
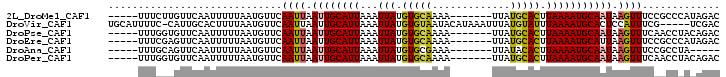
>2L_DroMel_CAF1 16089093 90 + 22407834 -----UUUCUUGUUCAAUUUUUAAUGUUCAAUUAAUUGCAUUAAAUUAUGUGCAAAA-------UUAUGCACUUAAAAUGCAAUAAGUUUCCGCCCAUAGAC -----..(((((..(......((((.....))))((((((((...(((.(((((...-------...))))).)))))))))))........)..)).))). ( -14.60) >DroVir_CAF1 27626 96 + 1 UGCAUUUUC-CAUUGCACUUUUAAUGUUCAAUUAAUUGCAUUAAAUUAUGUGUAAUACAUAAAUUUAUGUAUUUAAAAUGCACUCCAUUUCG-----UCGAC ((((((((.-..((((((.((((((((..........))))))))....))))))((((((....))))))...))))))))..........-----..... ( -20.20) >DroPse_CAF1 23596 90 + 1 -----UUUGGUGUUCAAUUUUUAAUGUUCAAUUAAUUGCAUUAAAUUAUGUGCAAAA-------UUAUGCACUUAAAAUGCAAUAAGUUUCAACCUACAGAC -----...(((....(((((.((((.....))))((((((((...(((.(((((...-------...))))).))))))))))))))))...)))....... ( -16.00) >DroEre_CAF1 19661 90 + 1 -----UUUCGAGUUCAAUUUUUAAUGUUCAAUUAAUUGCAUUAAAUUAUGUGCAAAA-------UUAUGCACUUAAAAUGCAUUAAGUUUCCGCCCAUAGAC -----..............((((((((..........))))))))(((((.((.(((-------(((((((.......)))))..)))))..)).))))).. ( -13.30) >DroAna_CAF1 17768 85 + 1 -----UUUGCAGUUCAAUUUUUAAUGUUCAAUUAAUUGCAUUAAAUUAUGUGCGAAA-------UUAUACACUUAAAAUGCAAUAAGUUUCCGCCUA----- -----.(((((.......(((((((((..((((..((((((........))))))))-------))..))).)))))))))))..............----- ( -11.71) >DroPer_CAF1 25588 90 + 1 -----UUUGGUGUUCAAUUUUUAAUGUUCAAUUAAUUGCAUUAAAUUAUGUGCAAAA-------UUAUGCACUUAAAAUGCAAUAAGUUUCAACCUACAGAC -----...(((....(((((.((((.....))))((((((((...(((.(((((...-------...))))).))))))))))))))))...)))....... ( -16.00) >consensus _____UUUCGUGUUCAAUUUUUAAUGUUCAAUUAAUUGCAUUAAAUUAUGUGCAAAA_______UUAUGCACUUAAAAUGCAAUAAGUUUCCGCCUAUAGAC .............................((((.((((((((...(((.(((((.............))))).))))))))))).))))............. ( -9.72 = -9.72 + 0.00)

| Location | 16,089,113 – 16,089,217 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -15.41 |
| Energy contribution | -15.80 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
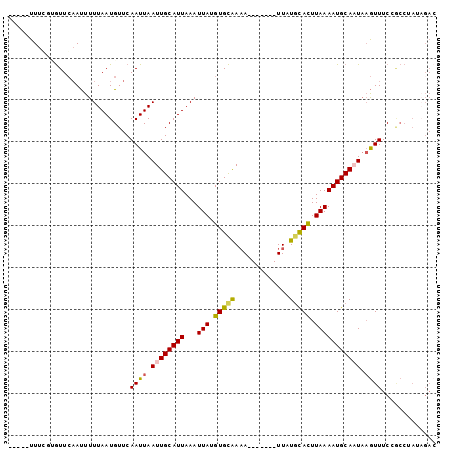
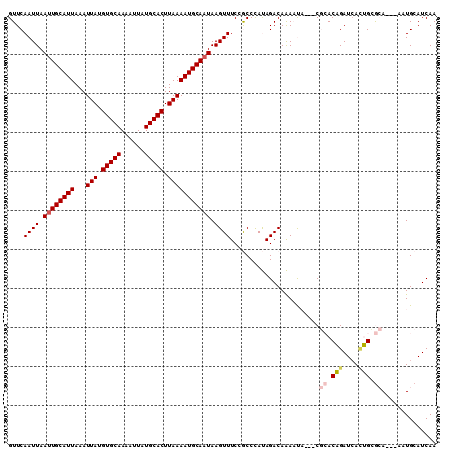
>2L_DroMel_CAF1 16089113 104 + 22407834 GUUCAAUUAAUUGCAUUAAAUUAUGUGCAAAAUUAUGCACUUAAAAUGCAAUAAGUUUCCGCCCAUAGACAAAAUA---CGCACAGAUCAGUGCGCA---AAUGCAUCAA ...........((((((...(((.(((((......))))).)))..(((.....((((........))))......---.((((......)))))))---)))))).... ( -21.90) >DroPse_CAF1 23616 107 + 1 GUUCAAUUAAUUGCAUUAAAUUAUGUGCAAAAUUAUGCACUUAAAAUGCAAUAAGUUUCAACCUACAGACAGACCUGACCCAACGGAUAAUCGGAAA---UCUGAAUCAA ((((((((.((((((((...(((.(((((......))))).))))))))))).))))........(((......))).......))))..((((...---.))))..... ( -21.00) >DroSec_CAF1 18127 107 + 1 GUUCAAUUAAUUGCAUUAAAUUAUGUGCAAAAUUAUGCACUUAAAAUGCAAUAAGUUUCCGCCCAUAGACAAAAUA---CGCACAGAUCACUGCGCAUAAAAUGCAUCAA ....((((.((((((((...(((.(((((......))))).))))))))))).))))...................---.((.(((....))).)).............. ( -19.80) >DroSim_CAF1 18138 107 + 1 GUUCAAUUAAUUGCAUUAAAUUAUGUGCAAAAUUAUGCACUUAAAAUGCAAUAAGUUUCCGCCCAUAGACAAAAUA---CGCACAGAUCACUGCGCAUAAAAUGCAUCAA ....((((.((((((((...(((.(((((......))))).))))))))))).))))...................---.((.(((....))).)).............. ( -19.80) >DroEre_CAF1 19681 104 + 1 GUUCAAUUAAUUGCAUUAAAUUAUGUGCAAAAUUAUGCACUUAAAAUGCAUUAAGUUUCCGCCCAUAGACAAAAUA---CGCACAGAUCACUGAGCA---AUUGCAUCAA (((((.(((((.(((((...(((.(((((......))))).)))))))))))))((((........))))......---............))))).---.......... ( -17.70) >DroPer_CAF1 25608 107 + 1 GUUCAAUUAAUUGCAUUAAAUUAUGUGCAAAAUUAUGCACUUAAAAUGCAAUAAGUUUCAACCUACAGACAGACCUGACCCAACGGAUCAUCGGAAA---UCUGAAUCAA (((.((((.((((((((...(((.(((((......))))).))))))))))).))))..)))...((((....(((((((....)).)))..))...---))))...... ( -21.20) >consensus GUUCAAUUAAUUGCAUUAAAUUAUGUGCAAAAUUAUGCACUUAAAAUGCAAUAAGUUUCCGCCCAUAGACAAAAUA___CGCACAGAUCACUGCGCA___AAUGCAUCAA ....((((.((((((((...(((.(((((......))))).))))))))))).)))).......................((.(((....))).)).............. (-15.41 = -15.80 + 0.39)


| Location | 16,089,145 – 16,089,235 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -20.33 |
| Energy contribution | -20.83 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
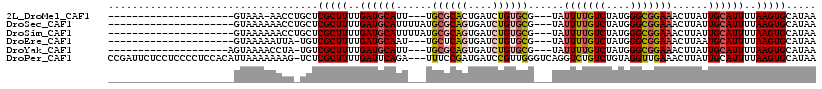
>2L_DroMel_CAF1 16089145 90 - 22407834 ---------------------GUAAA-AACCUGCUCGCUUUUGAUGCAUU---UGCGCACUGAUCUGUGCG---UAUUUUGUCUAUGGGCGGAAACUUAUUGCAUUUUAAGUGCAUAA ---------------------.....-....(((..((((..((((((..---(((((((......)))))---))(((((((....)))))))......))))))..)))))))... ( -27.90) >DroSec_CAF1 18159 94 - 1 ---------------------GUAAAAAACCUGCUCGCUUUUGAUGCAUUUUAUGCGCAGUGAUCUGUGCG---UAUUUUGUCUAUGGGCGGAAACUUAUUGCAUUUUAAGUGCAUAA ---------------------..........(((..((((..((((((...(((((((((....)))))))---))(((((((....)))))))......))))))..)))))))... ( -29.20) >DroSim_CAF1 18170 94 - 1 ---------------------GUAAAAAACCUGCUCGCUUUUGAUGCAUUUUAUGCGCAGUGAUCUGUGCG---UAUUUUGUCUAUGGGCGGAAACUUAUUGCAUUUUAAGUGCAUAA ---------------------..........(((..((((..((((((...(((((((((....)))))))---))(((((((....)))))))......))))))..)))))))... ( -29.20) >DroEre_CAF1 19713 90 - 1 ---------------------GUAAAAAUUA-UGUCGCUUUUGAUGCAAU---UGCUCAGUGAUCUGUGCG---UAUUUUGUCUAUGGGCGGAAACUUAAUGCAUUUUAAGUGCAUAA ---------------------.......(((-((.(((((..((((((..---.((.(((....))).))(---(.(((((((....)))))))))....))))))..)))))))))) ( -27.20) >DroYak_CAF1 19181 91 - 1 --------------------AGUAAAACCUA-UGUCGCUUUUGAUGCAUU---UGCGCAGUGAUCUGUGCG---UAUUUUGUCUAUGGGCGGAAACUUAUUGCAUUUUAAGUGCAUAA --------------------.........((-((.(((((..((((((..---.((((((....))))))(---(.(((((((....)))))))))....))))))..))))))))). ( -30.20) >DroPer_CAF1 25640 114 - 1 CCGAUUCUCCUCCCCUCCACAUUAAAAAAAG-UCUCGCUUUUGAUUCAGA---UUUCCGAUGAUCCGUUGGGUCAGGUCUGUCUGUAGGUUGAAACUUAUUGCAUUUUAAGUGCAUAA ....(((.(((...............(((((-.....)))))....((((---(..((..((((((...))))))))...))))).)))..)))......(((((.....)))))... ( -21.60) >consensus _____________________GUAAAAAACC_GCUCGCUUUUGAUGCAUU___UGCGCAGUGAUCUGUGCG___UAUUUUGUCUAUGGGCGGAAACUUAUUGCAUUUUAAGUGCAUAA ...................................(((((..((((((......((((((....))))))......(((((((....)))))))......))))))..)))))..... (-20.33 = -20.83 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:43 2006