
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,086,826 – 16,086,946 |
| Length | 120 |
| Max. P | 0.683056 |

| Location | 16,086,826 – 16,086,946 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.92 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -20.67 |
| Energy contribution | -20.37 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16086826 120 + 22407834 CAGGAGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAAACUUGCAGCGAAAGCGCGCUUA ...((((............((((.....((((((....)))))).....)))).........((...((((..(((((((.......)))))))....))))...((....)))))))). ( -32.60) >DroPse_CAF1 19883 120 + 1 CAGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGAAAGUCUAAAACGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAAAGUUUCAGUGAAGGCGCGAGGA ..((...............((((.....((((((....)))))).....)))).....(((((((.(((.(..(((((((.......)))))))....).))).)))..))))))..... ( -27.00) >DroGri_CAF1 17093 112 + 1 UGGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAGAAUGUCUGCUUAAACUGUUCUAAGUGAUUGUAAUUUAGGA-UAAGUUGCGCGUAA-------CUC ..(((((............((((.....((((((....)))))).....)))).................((((((((((.......))))))))-)).)))))......-------... ( -29.50) >DroYak_CAF1 16881 109 + 1 CAGCAGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAAACUUG-----------CGCUAA .((((((((.((.......((((.....((((((....)))))).....)))))).))).)))))...((((((((((((.......)))))))........)-----------)))).. ( -26.51) >DroMoj_CAF1 26323 112 + 1 UGGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAACUGUUUUAAGUGAUUGUAAUUUAGGA-UAAGUUGCGAGCAC-------CUC ((....))...........((((.....((((((....)))))).....))))........((((..(((((((((((((.......))))))))-).))))...)))).-------... ( -28.10) >DroPer_CAF1 22271 120 + 1 CAGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAACGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAAAGUUUCAGUGAAGGCGCGAGGA ..((...............((((.....((((((....)))))).....)))).....(((((((.(((.(..(((((((.......)))))))....).))).)))..))))))..... ( -27.00) >consensus CAGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAAAGUUGCAGGGAA____CG_CUA .((((((((.((....)).((((.....((((((....)))))).....))))...)))).))))..((((..(((((((.......)))))))....)))).................. (-20.67 = -20.37 + -0.30)

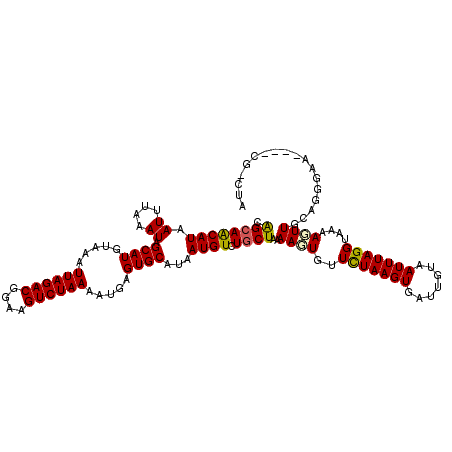
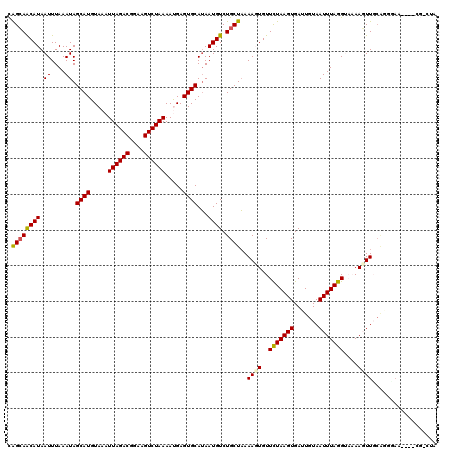
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:41 2006