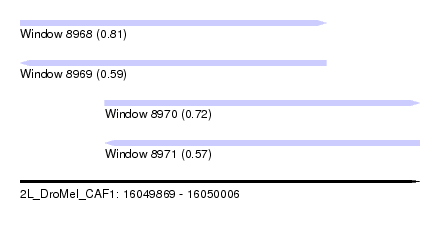
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,049,869 – 16,050,006 |
| Length | 137 |
| Max. P | 0.808201 |

| Location | 16,049,869 – 16,049,974 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.44 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -25.12 |
| Energy contribution | -26.07 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
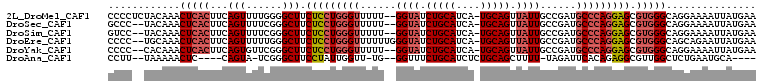
>2L_DroMel_CAF1 16049869 105 + 22407834 CCCCUCUACAAACUCACUUCAGUUUUGGGGCUUCUCCUGGGUUUUU--GGUAUCUGCAUCA-UGCAGUUAUUGCCGAUGCCCAGGAGCGUGGGCAGGAAAAUUAUGAA ((((((((.(((((......))))))))))...(((((((((..((--((((.((((....-.))))....)))))).)))))))))...)))............... ( -38.10) >DroSec_CAF1 225423 103 + 1 GCCC--UACAAACUCACUUCAGUUUUCGGGCUUCUCCUGGGUUUUU--GGUAUCUGCAUCA-UGCAGUUAUUGCCGAUGCCCAGGAGCGUGGGCAGGAAAAUUAUGAA ((((--.(((((((......)))))........(((((((((..((--((((.((((....-.))))....)))))).))))))))).)))))).............. ( -39.90) >DroSim_CAF1 232236 103 + 1 GUCC--UACAAACUCACUUCAGUUUUCGGGCUUCUCCUGGGUUUUU--GGUAUCUGCAUCA-UGCAGUUAUUGCCGAUGCCCAGGAGCGUGGGCAGGAAAAUUAUGAA ....--............(((((((((..(((((((((((((..((--((((.((((....-.))))....)))))).)))))))))...))))..))))))..))). ( -39.10) >DroEre_CAF1 214322 105 + 1 CCCC--UGCAAACUCACUUCAGUUUUUGGGCUUCUCCUGGGUUUUUUGGGUAUCUGCAUCA-UGCAGUUAUUGCCGAUGCCCAGGAGCGUGGGCAGCAGAAUUAUGAA ...(--(((...(((((...((((....)))).(((((((((....(.((((.((((....-.))))....)))).).))))))))).)))))..))))......... ( -38.80) >DroYak_CAF1 211614 103 + 1 CCCC--CACAAACUCACUUCAGUGUUCGGGCUUCUCCUGGGUUUUU--GGUAUCUGCAUCA-UGCAGUUAUUGCCGAUGCCCAGGAGCGUGGGCAGGAAAAUUAUGAA ((((--(((.....(((....))).........(((((((((..((--((((.((((....-.))))....)))))).))))))))).)))))..))........... ( -40.60) >DroAna_CAF1 158278 93 + 1 CCUU--UAAAAACUC----CAGUA-UCGGGCUUCCUAUUGGUU-UG--GGUUUCUGCAUCUCUGCAGCUUUU-UAGAUUCACAGAGGCGUUGGCUCUGAAUGCA---- ....--.........----..(((-(((((((.(((..(((((-((--((...(((((....)))))...))-)))).)))...)))....))).))).)))).---- ( -20.60) >consensus CCCC__UACAAACUCACUUCAGUUUUCGGGCUUCUCCUGGGUUUUU__GGUAUCUGCAUCA_UGCAGUUAUUGCCGAUGCCCAGGAGCGUGGGCAGGAAAAUUAUGAA ............(((((...(((......))).(((((((((......((((.(((((....))))).))))......))))))))).)))))............... (-25.12 = -26.07 + 0.95)
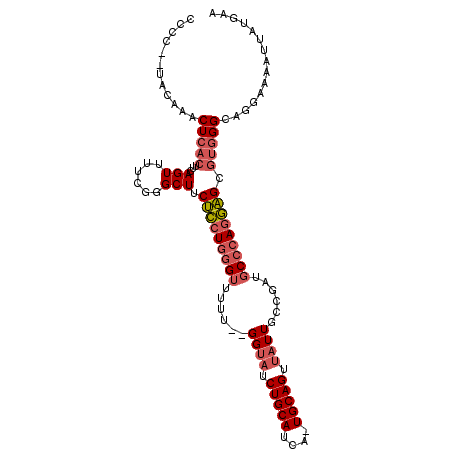

| Location | 16,049,869 – 16,049,974 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.44 |
| Mean single sequence MFE | -29.61 |
| Consensus MFE | -21.64 |
| Energy contribution | -22.58 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16049869 105 - 22407834 UUCAUAAUUUUCCUGCCCACGCUCCUGGGCAUCGGCAAUAACUGCA-UGAUGCAGAUACC--AAAAACCCAGGAGAAGCCCCAAAACUGAAGUGAGUUUGUAGAGGGG .....................((((((((((((((((.....))).-)))))).(....)--......)))))))...((((.(((((......))))).....)))) ( -30.10) >DroSec_CAF1 225423 103 - 1 UUCAUAAUUUUCCUGCCCACGCUCCUGGGCAUCGGCAAUAACUGCA-UGAUGCAGAUACC--AAAAACCCAGGAGAAGCCCGAAAACUGAAGUGAGUUUGUA--GGGC .....................((((((((((((((((.....))).-)))))).(....)--......)))))))..((((..(((((......)))))...--)))) ( -33.00) >DroSim_CAF1 232236 103 - 1 UUCAUAAUUUUCCUGCCCACGCUCCUGGGCAUCGGCAAUAACUGCA-UGAUGCAGAUACC--AAAAACCCAGGAGAAGCCCGAAAACUGAAGUGAGUUUGUA--GGAC ..........((((((.....((((((((((((((((.....))).-)))))).(....)--......)))))))........(((((......))))))))--))). ( -31.80) >DroEre_CAF1 214322 105 - 1 UUCAUAAUUCUGCUGCCCACGCUCCUGGGCAUCGGCAAUAACUGCA-UGAUGCAGAUACCCAAAAAACCCAGGAGAAGCCCAAAAACUGAAGUGAGUUUGCA--GGGG .......((((((.((.(((.((((((((((((((((.....))).-)))))).(.....).......))))))).....((.....))..))).))..)))--))). ( -32.60) >DroYak_CAF1 211614 103 - 1 UUCAUAAUUUUCCUGCCCACGCUCCUGGGCAUCGGCAAUAACUGCA-UGAUGCAGAUACC--AAAAACCCAGGAGAAGCCCGAACACUGAAGUGAGUUUGUG--GGGG ...............(((((.((((((((((((((((.....))).-)))))).(....)--......))))))).........(((....))).....)))--)).. ( -33.70) >DroAna_CAF1 158278 93 - 1 ----UGCAUUCAGAGCCAACGCCUCUGUGAAUCUA-AAAAGCUGCAGAGAUGCAGAAACC--CA-AACCAAUAGGAAGCCCGA-UACUG----GAGUUUUUA--AAGG ----.((((.....((....))((((((.......-.......))))))))))(((((((--((-........((....))..-...))----).)))))).--.... ( -16.46) >consensus UUCAUAAUUUUCCUGCCCACGCUCCUGGGCAUCGGCAAUAACUGCA_UGAUGCAGAUACC__AAAAACCCAGGAGAAGCCCGAAAACUGAAGUGAGUUUGUA__GGGG ...............(((...((((((((............(((((....)))))............))))))))........(((((......))))).....))). (-21.64 = -22.58 + 0.95)

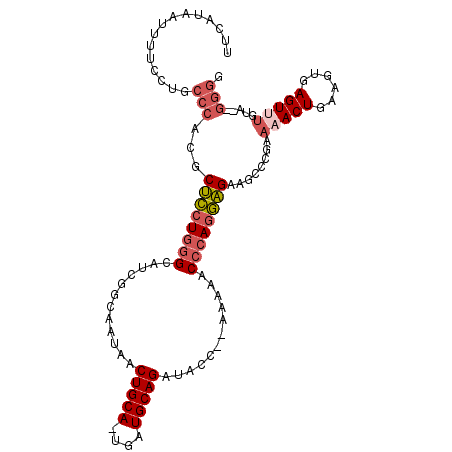
| Location | 16,049,898 – 16,050,006 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.00 |
| Mean single sequence MFE | -39.07 |
| Consensus MFE | -24.35 |
| Energy contribution | -24.77 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16049898 108 + 22407834 GCUUCUCCUGGGUUUUU--GGUAUCUGCAUC-AUGCAGUUAUUGCCGAUGCCCAGGAGCGUGGGCAGGAAAAUUAUGAAGGAGAAUA--UCG--GGGGCACCUACC----ACGCCGCUU ((...((((((((..((--((((.((((...-..))))....)))))).))))))))((((((.................((.....--))(--((....))).))----)))).)).. ( -42.80) >DroPse_CAF1 260585 107 + 1 -CUCCUCCUGUGUGUGU--GGUAUCUGCAUCUAUGCAGUUAUUGUAUGUGGCAAAGGGCGUAGGCAUUACGAUUAUCGAGGGA------UCGUACGAGUACCUGACUUUUAAAAUG--- -....((((.((((((.--.(((.(((((....))))).)))..)))...))).))))..((((.(((((((((.......))------)))))...)).))))............--- ( -25.10) >DroSec_CAF1 225450 108 + 1 GCUUCUCCUGGGUUUUU--GGUAUCUGCAUC-AUGCAGUUAUUGCCGAUGCCCAGGAGCGUGGGCAGGAAAAUUAUGAAGGAGAAUA--UCG--GGGGCCACUACC----ACGGCGCUU ((..(((((((((..((--((((.((((...-..))))....)))))).)))))))))(((((...((.....(((........)))--...--....))....))----))))).... ( -40.10) >DroSim_CAF1 232263 108 + 1 GCUUCUCCUGGGUUUUU--GGUAUCUGCAUC-AUGCAGUUAUUGCCGAUGCCCAGGAGCGUGGGCAGGAAAAUUAUGAAGGAGAAUA--UCG--GGGGCCACUACC----ACGGCGUUU ((..(((((((((..((--((((.((((...-..))))....)))))).)))))))))(((((...((.....(((........)))--...--....))....))----))))).... ( -40.10) >DroEre_CAF1 214349 109 + 1 GCUUCUCCUGGGUUUUUUGGGUAUCUGCAUC-AUGCAGUUAUUGCCGAUGCCCAGGAGCGUGGGCAGCAGAAUUAUGAAGGGGA-UA--UCG--GGGGUCCCCACC----AAAGCGCUU (((((((((((((....(.((((.((((...-..))))....)))).).)))))))))..(((....((......))..(((((-(.--...--...)))))).))----))))).... ( -43.20) >DroYak_CAF1 211641 110 + 1 GCUUCUCCUGGGUUUUU--GGUAUCUGCAUC-AUGCAGUUAUUGCCGAUGCCCAGGAGCGUGGGCAGGAAAAUUAUGAAGGGGAAUAUUCUG--GGGGUCCCUACC----AUACUGCUU ....(((((((((..((--((((.((((...-..))))....)))))).)))))))))...((((((......((((..(((((...((...--.)).)))))..)----))))))))) ( -43.10) >consensus GCUUCUCCUGGGUUUUU__GGUAUCUGCAUC_AUGCAGUUAUUGCCGAUGCCCAGGAGCGUGGGCAGGAAAAUUAUGAAGGAGAAUA__UCG__GGGGCCCCUACC____ACAGCGCUU (((((((((((((......((((.(((((....))))).))))......))))))))((....)).............................))))).................... (-24.35 = -24.77 + 0.42)

| Location | 16,049,898 – 16,050,006 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.00 |
| Mean single sequence MFE | -30.19 |
| Consensus MFE | -19.66 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16049898 108 - 22407834 AAGCGGCGU----GGUAGGUGCCCC--CGA--UAUUCUCCUUCAUAAUUUUCCUGCCCACGCUCCUGGGCAUCGGCAAUAACUGCAU-GAUGCAGAUACC--AAAAACCCAGGAGAAGC ..((.((((----((((((......--.((--.........))........)))).))))))(((((((((((((((.....))).)-))))).(....)--......))))))...)) ( -34.16) >DroPse_CAF1 260585 107 - 1 ---CAUUUUAAAAGUCAGGUACUCGUACGA------UCCCUCGAUAAUCGUAAUGCCUACGCCCUUUGCCACAUACAAUAACUGCAUAGAUGCAGAUACC--ACACACACAGGAGGAG- ---..........((.(((((....(((((------(.........)))))).)))))))..(((((..............(((((....))))).....--.........)))))..- ( -19.89) >DroSec_CAF1 225450 108 - 1 AAGCGCCGU----GGUAGUGGCCCC--CGA--UAUUCUCCUUCAUAAUUUUCCUGCCCACGCUCCUGGGCAUCGGCAAUAACUGCAU-GAUGCAGAUACC--AAAAACCCAGGAGAAGC ..((..(((----(((((.((...)--)((--.........)).........))).)))))((((((((((((((((.....))).)-))))).(....)--......)))))))..)) ( -30.30) >DroSim_CAF1 232263 108 - 1 AAACGCCGU----GGUAGUGGCCCC--CGA--UAUUCUCCUUCAUAAUUUUCCUGCCCACGCUCCUGGGCAUCGGCAAUAACUGCAU-GAUGCAGAUACC--AAAAACCCAGGAGAAGC ....(((((----(((((.((...)--)((--.........)).........))).)))))((((((((((((((((.....))).)-))))).(....)--......)))))))..)) ( -29.50) >DroEre_CAF1 214349 109 - 1 AAGCGCUUU----GGUGGGGACCCC--CGA--UA-UCCCCUUCAUAAUUCUGCUGCCCACGCUCCUGGGCAUCGGCAAUAACUGCAU-GAUGCAGAUACCCAAAAAACCCAGGAGAAGC .((((...(----((.(((((....--...--..-))))).)))......)))).......((((((((((((((((.....))).)-))))).(.....).......))))))).... ( -33.50) >DroYak_CAF1 211641 110 - 1 AAGCAGUAU----GGUAGGGACCCC--CAGAAUAUUCCCCUUCAUAAUUUUCCUGCCCACGCUCCUGGGCAUCGGCAAUAACUGCAU-GAUGCAGAUACC--AAAAACCCAGGAGAAGC ..((.((..----((((((((....--..(((........)))......)))))))).)).((((((((((((((((.....))).)-))))).(....)--......)))))))..)) ( -33.80) >consensus AAGCGCCGU____GGUAGGGACCCC__CGA__UAUUCCCCUUCAUAAUUUUCCUGCCCACGCUCCUGGGCAUCGGCAAUAACUGCAU_GAUGCAGAUACC__AAAAACCCAGGAGAAGC .............(((((..................................)))))....((((((((............(((((....)))))............)))))))).... (-19.66 = -20.25 + 0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:34 2006