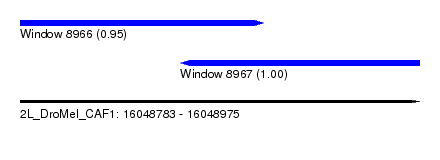
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,048,783 – 16,048,975 |
| Length | 192 |
| Max. P | 0.998052 |

| Location | 16,048,783 – 16,048,900 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -29.54 |
| Consensus MFE | -23.84 |
| Energy contribution | -24.56 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16048783 117 + 22407834 UUGUGCUAAUCCUAAAGGAAUUUGCUC---GCAAAUUUAUGGCUUCGUUAUCCCACUUGAGCUUUGAUCUACAAAAGCUGGUUAAAUGGCCAAGGAUUUAUUCAAUUAGUAUUGGCAUAA .(((((((((.((((..(((((((...---.))))))).(((((...(((.((......((((((........)))))))).)))..))))).............)))).))))))))). ( -31.70) >DroSec_CAF1 224371 117 + 1 UUGUGCUAAUCCUAAAGGAAUUUGCUC---GCAAAUUUAUGGCUUCGUUAUCCCACUUGAGCUUUGAUCUACAAAAGCUGGUUAAAUGGCCAAGGAUUUAUUCAAUUAGUAUUGGCAUAA .(((((((((.((((..(((((((...---.))))))).(((((...(((.((......((((((........)))))))).)))..))))).............)))).))))))))). ( -31.70) >DroSim_CAF1 231157 117 + 1 UUGUGCUAAUCCUAAAGGAAUUUGCUC---GCAAAUUUAUGGCUUCGUUAUCCCACUUGAGCUUUGAUCUACAAAAGCUGGUUAAAUGGCCAAGGAUUUAUUCAAUUAGUAUUGGCAUAA .(((((((((.((((..(((((((...---.))))))).(((((...(((.((......((((((........)))))))).)))..))))).............)))).))))))))). ( -31.70) >DroEre_CAF1 213246 120 + 1 UUCUGCUGGUCCGAAAGGAAUUUCCAAGAAGGAAAUAUAUGGCUUUGUCAUCUCACUUGAGCUUUGAUCUACAAAAGCUGGUAAAAUGACCAAGGAUUUCUUCAAUUAGUAUUGGCAUAA ...(((..((((....)))((((((.....))))))......(((.(((((.(.(((..((((((........))))))))).).))))).))).................)..)))... ( -28.90) >DroYak_CAF1 210526 110 + 1 UUCUGCUAGUCCGAAAGGAAUUUUCUG---GCAAAUACAUGGCUUUGUUAUCCCGCUUGAGCUU-------CAAAAACUGGUGAAAUGGCCAAGGAUUUAUUCAAUUAGUUUUGGCAUAA ...(((((((((....))).....)))---)))....(.(((((...(((((....(((.....-------))).....)))))...))))).)...((((((((......)))).)))) ( -23.70) >consensus UUGUGCUAAUCCUAAAGGAAUUUGCUC___GCAAAUUUAUGGCUUCGUUAUCCCACUUGAGCUUUGAUCUACAAAAGCUGGUUAAAUGGCCAAGGAUUUAUUCAAUUAGUAUUGGCAUAA .(((((((((.((((..((((((((.....))))))))....(((.(((((...(((..((((((........)))))))))...))))).)))...........)))).))))))))). (-23.84 = -24.56 + 0.72)

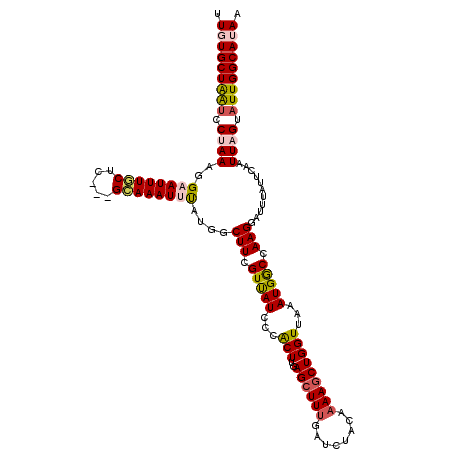
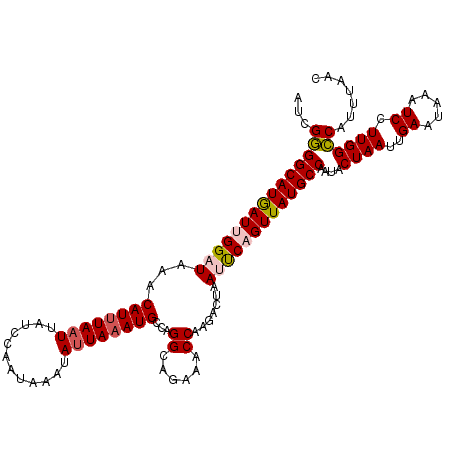
| Location | 16,048,860 – 16,048,975 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -20.60 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16048860 115 - 22407834 AUCGGGGCAUGAUUGGAUAAACAUCUAUUUAUGCAAUAAAUAUUAAAUGCAAGGCAGAAACCAAGACUAAUUCAGUUAUGCCAAUACUAAUUGAAUAAAUCCUUGGCCAUUUAAC ...((.((((((.(((((....))))).))))))..........................(((((....(((((((((.(......)))))))))).....)))))))....... ( -23.30) >DroSec_CAF1 224448 115 - 1 AUCGGGGCAUGAUUGGAUAAACAUUUAAUUAUCCAAUAAAUAUUAAAUGCCAGGCAGAAACCAAGUCUAAUUCAGUUAUGCCAAUACUAAUUGAAUAAAUCCUUGGCCAUUUAAC ...(((((((.(((((((((........))))))))).........))))).........(((((....(((((((((.(......)))))))))).....)))))))....... ( -27.30) >DroSim_CAF1 231234 115 - 1 AUCGGGGCAUGAUUGGAUAAACAUUUAAUUAUCCAAUAAAUAUUAAAUGCCAGGCAGAAACCAAGACUAAUUCAGUUAUGCCAAUACUAAUUGAAUAAAUCCUUGGCCAUUUAAC ...(((((((.(((((((((........))))))))).........))))).........(((((....(((((((((.(......)))))))))).....)))))))....... ( -27.30) >DroEre_CAF1 213326 114 - 1 UUCGAGGCAUAAUGGGAUAAACAUUUAAUUAUCCAAUAAAUAUUAAAUGCCCGGGAGA-ACCAAGACUAACCCAGUUAUGCCAAUACUAAUUGAAGAAAUCCUUGGUCAUUUUAC .....(((((((((((.....((((((((............))))))))...((....-.))........)))).)))))))...(((((..((.....)).)))))........ ( -22.50) >DroYak_CAF1 210596 115 - 1 UUCGGGGCAUAAUAGGAUAAGCAUUUAAUUAUCCAAUAAAUAUUAAAUGCCAGGCAGAAACCAAUACUAACCCAGUUAUGCCAAAACUAAUUGAAUAAAUCCUUGGCCAUUUCAC ...((((((((((.((....(((((((((............)))))))))..((......)).........)).))))))))....((((..((.....)).))))))....... ( -23.10) >consensus AUCGGGGCAUGAUUGGAUAAACAUUUAAUUAUCCAAUAAAUAUUAAAUGCCAGGCAGAAACCAAGACUAAUUCAGUUAUGCCAAUACUAAUUGAAUAAAUCCUUGGCCAUUUAAC ...(((((((((((((((...((((((((............))))))))...((......)).......)))))))))))))....((((..((.....)).))))))....... (-20.60 = -21.00 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:30 2006