
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,022,188 – 16,022,286 |
| Length | 98 |
| Max. P | 0.540641 |

| Location | 16,022,188 – 16,022,286 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -20.21 |
| Energy contribution | -20.27 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16022188 98 + 22407834 AGUUGCAACUGGCCAUGCAAGAUUGCUGGCG---AA---------------AAACAAAAG----GUAUAUUUGUAUCUAUGCAUUUUGUAUGCGGGUUUUCAGCAACUGUUGUCCACCAG ((((((.....((((.(((....)))))))(---((---------------((.(...((----((((....)))))).(((((.....)))))).))))).))))))............ ( -27.20) >DroPse_CAF1 222817 115 + 1 AAUUGCAGCUGGCCAUGCAAGAUUUCUGGUAACGGAAACCGAGCGGGGAA-AGGCAAAAA----GUAUAUUUGUAUCUAUGCAUUUUGCAUGCGGGCUUUCAGCAACUGUUGUCUGUCAG ...((((.((..((.(((..(.((((((....)))))).)..))).))..-))((((((.----(((((........))))).)))))).))))(((...((((....))))...))).. ( -33.30) >DroEre_CAF1 186843 98 + 1 AGUUGCAGCUGGACAUGCAAGAUUGCUGGCG---AA---------------AAGCAAAAA----GUACAUUUGUAUCUAUGCAUUUUGUAUGCGGGUUUUCAGCAACUGUUGUCCACCAG ((((((((((.(.((((((((((.((..((.---..---------------..)).....----((((....))))....))))))))))))).))))....))))))............ ( -26.30) >DroYak_CAF1 182872 98 + 1 AGUUGCAGUUGGUCAUGCAAGAUAGCUGACG---GA---------------AAAUAAAAG----GUAUAUUUGUAUCUAUGCAUUUUGUAUGCGGGCUUUCAGCAACUGUUGUCCACCAG ((((((((((.(.((((((((((.((.((..---..---------------(((((....----...)))))...))...))))))))))))).))))....))))))............ ( -23.60) >DroAna_CAF1 131391 120 + 1 AGUUGCAGCUGGGCAGGCAAGAUUGCUGGUGUCGAAAACUGCGAAGGGAAUAAACAAAAAGUCCGUAUAUUUGUAUCUAUGCAUUUUGCAUGCGGGCUUUCAGCAACUGUUGUCGCUCAG ........((((((.(((((.(((((((...(((.......))).............(((((((((((...((((....))))......))))))))))))))))).).))))))))))) ( -36.60) >DroPer_CAF1 252303 115 + 1 AAUUGCAGCUGGCCAUGCAAGAUUUCUGGUAACGGAAACCGAGCGGGGAA-AGGCAAAAA----GUAUAUUUGUAUCUAUGCAUUUUGCAUGCGGGCUUUCAGCAACUGUUGUCUGUCAG ...((((.((..((.(((..(.((((((....)))))).)..))).))..-))((((((.----(((((........))))).)))))).))))(((...((((....))))...))).. ( -33.30) >consensus AGUUGCAGCUGGCCAUGCAAGAUUGCUGGCG___AA_______________AAACAAAAA____GUAUAUUUGUAUCUAUGCAUUUUGCAUGCGGGCUUUCAGCAACUGUUGUCCACCAG ((((((((((.(.((((((((((...............((....))..................(((((........)))))))))))))))).))))....))))))............ (-20.21 = -20.27 + 0.06)

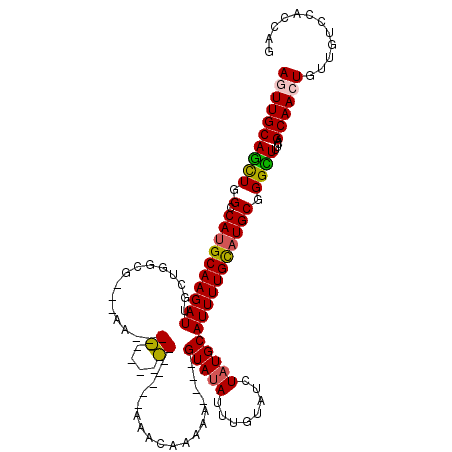
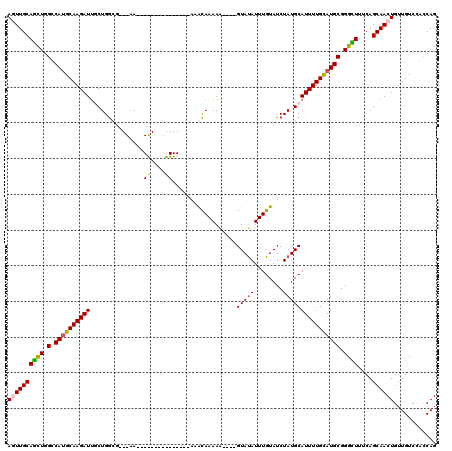
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:19 2006