| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,018,753 – 16,018,853 |
| Length | 100 |
| Max. P | 0.858299 |

| Location | 16,018,753 – 16,018,853 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.15 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -19.07 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
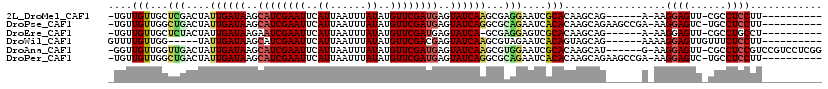
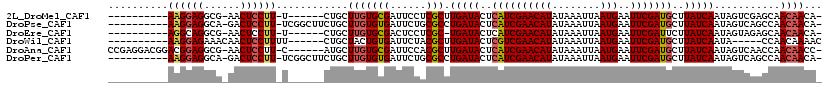
>2L_DroMel_CAF1 16018753 100 + 22407834 -UGUUGUUGCUCGACUAUUGAUAAGCAUCGAAUUCAUUAAUUUAUAUGUUCGAUGAGUAUCAAGCGAGGAAUCGCACAAGCAG------A-AAGGAGUU-CGCCUCCUU---------- -(((.(((.((((....((((((..((((((((..((......))..))))))))..)))))).)))).))).))).......------.-((((((..-...))))))---------- ( -32.90) >DroPse_CAF1 218951 106 + 1 -UGUUGUUGGCUGACUAUUGAUAAGCAUCGAAUUCAUUAAUUUAUAUGUUCGAUGAGUAUCAGGCGCAGAAUCACACAAGCAGAAGCCGA-AAGGAGUC-UGCCUCCUU---------- -..((((((.(((....((((((..((((((((..((......))..))))))))..))))))...)))...)).))))(((((..((..-..))..))-)))......---------- ( -36.00) >DroEre_CAF1 183365 99 + 1 -UGUUGUUGCUCUACUAUUGAUAAGAAUCGAAUUCAUUAAUUUAUAUGUUCGAUGAGUAUCA-GCGAGGAGUCGCACAAGCAG------A-AAGGAGUU-CGCCUGCCU---------- -(((.((.(((((....((((((...(((((((..((......))..)))))))...)))))-)...))))).))))).((((------.-........-...))))..---------- ( -23.60) >DroWil_CAF1 183679 98 + 1 GUUUUGUUGG-----UAUUGAUAAGCAUCGAAUUCAUUAAUUUAUAUGUUCGACGAGUAUCAAGCGUAGAAUCACAGUAGCAG------AAAAGGAGUUGUUUCUCCUU---------- .((((((((.-----(.((((((..(.((((((..((......))..)))))).)..))))))..((......))).))))))------))((((((......))))))---------- ( -23.50) >DroAna_CAF1 128155 110 + 1 -GGUUGUUGGUUGACUAUUGAUAAGCAUCGAAUUCAUUAAUUUAUAUGUUCGAUGAGUAUCAAGCGUGGAAUCGCACAAGCAU------G-AAGGAGUU-CGCCUCCGUCCGUCCUCGG -((.((..(((.((((.((((((..((((((((..((......))..))))))))..))))))(((......)))........------.-....))))-.)))..)).))........ ( -30.90) >DroPer_CAF1 248419 106 + 1 -UGUUGUUGGCUGACUAUUGAUAAGCAUCGAAUUCAUUAAUUUAUAUGUUCGAUGAGUAUCAGGCGCAGAAUCACACAAGCAGAAGCCGA-AAGGAGUC-UGCCUCCUU---------- -..((((((.(((....((((((..((((((((..((......))..))))))))..))))))...)))...)).))))(((((..((..-..))..))-)))......---------- ( -36.00) >consensus _UGUUGUUGGUUGACUAUUGAUAAGCAUCGAAUUCAUUAAUUUAUAUGUUCGAUGAGUAUCAAGCGAAGAAUCACACAAGCAG______A_AAGGAGUU_CGCCUCCUU__________ ....(((...(((....((((((..((((((((..((......))..))))))))..))))))...)))....))).................((((......))))............ (-19.07 = -19.27 + 0.19)
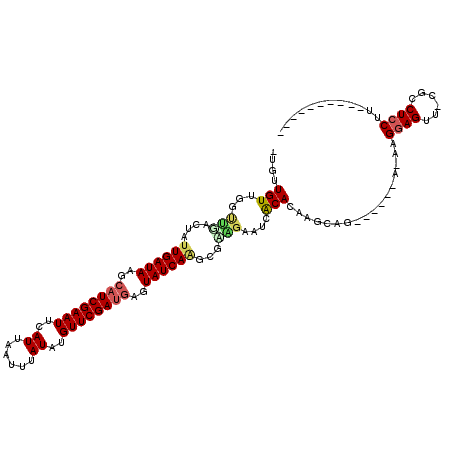
| Location | 16,018,753 – 16,018,853 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.15 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -18.65 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
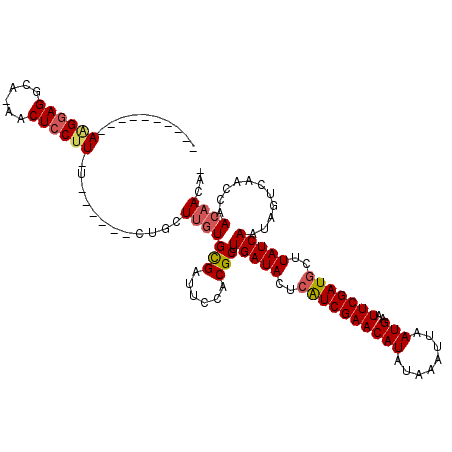
>2L_DroMel_CAF1 16018753 100 - 22407834 ----------AAGGAGGCG-AACUCCUU-U------CUGCUUGUGCGAUUCCUCGCUUGAUACUCAUCGAACAUAUAAAUUAAUGAAUUCGAUGCUUAUCAAUAGUCGAGCAACAACA- ----------((((((...-..))))))-.------.((((((.((((....))))((((((..((((((((((........)))..)))))))..))))))....))))))......- ( -30.60) >DroPse_CAF1 218951 106 - 1 ----------AAGGAGGCA-GACUCCUU-UCGGCUUCUGCUUGUGUGAUUCUGCGCCUGAUACUCAUCGAACAUAUAAAUUAAUGAAUUCGAUGCUUAUCAAUAGUCAGCCAACAACA- ----------((((((...-..))))))-..((((.(((...((((......)))).(((((..((((((((((........)))..)))))))..))))).)))..)))).......- ( -28.60) >DroEre_CAF1 183365 99 - 1 ----------AGGCAGGCG-AACUCCUU-U------CUGCUUGUGCGACUCCUCGC-UGAUACUCAUCGAACAUAUAAAUUAAUGAAUUCGAUUCUUAUCAAUAGUAGAGCAACAACA- ----------(((.((...-..)))))(-(------(((((...((((....))))-(((((...(((((((((........)))..))))))...)))))..)))))))........- ( -20.50) >DroWil_CAF1 183679 98 - 1 ----------AAGGAGAAACAACUCCUUUU------CUGCUACUGUGAUUCUACGCUUGAUACUCGUCGAACAUAUAAAUUAAUGAAUUCGAUGCUUAUCAAUA-----CCAACAAAAC ----------((((((......))))))..------........(((......)))((((((..((((((((((........)))..)))))))..))))))..-----.......... ( -19.00) >DroAna_CAF1 128155 110 - 1 CCGAGGACGGACGGAGGCG-AACUCCUU-C------AUGCUUGUGCGAUUCCACGCUUGAUACUCAUCGAACAUAUAAAUUAAUGAAUUCGAUGCUUAUCAAUAGUCAACCAACAACC- .....((((((.((((...-..))))..-.------.(((....)))..)))....((((((..((((((((((........)))..)))))))..))))))..)))...........- ( -27.30) >DroPer_CAF1 248419 106 - 1 ----------AAGGAGGCA-GACUCCUU-UCGGCUUCUGCUUGUGUGAUUCUGCGCCUGAUACUCAUCGAACAUAUAAAUUAAUGAAUUCGAUGCUUAUCAAUAGUCAGCCAACAACA- ----------((((((...-..))))))-..((((.(((...((((......)))).(((((..((((((((((........)))..)))))))..))))).)))..)))).......- ( -28.60) >consensus __________AAGGAGGCA_AACUCCUU_U______CUGCUUGUGCGAUUCCACGCUUGAUACUCAUCGAACAUAUAAAUUAAUGAAUUCGAUGCUUAUCAAUAGUCAACCAACAACA_ ..........((((((......))))))............(((((((......))).(((((..((((((((((........)))..)))))))..)))))...........))))... (-18.65 = -18.98 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:16 2006