| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,003,805 – 16,003,896 |
| Length | 91 |
| Max. P | 0.631299 |

| Location | 16,003,805 – 16,003,896 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -18.72 |
| Energy contribution | -19.47 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
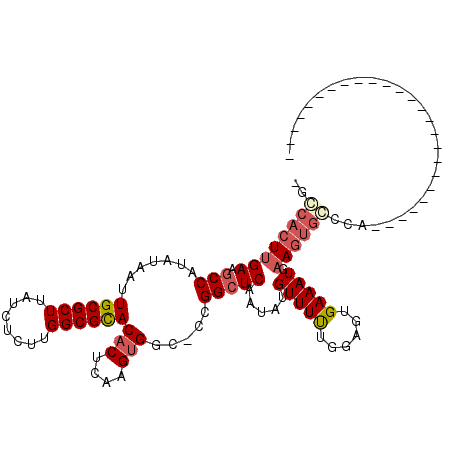
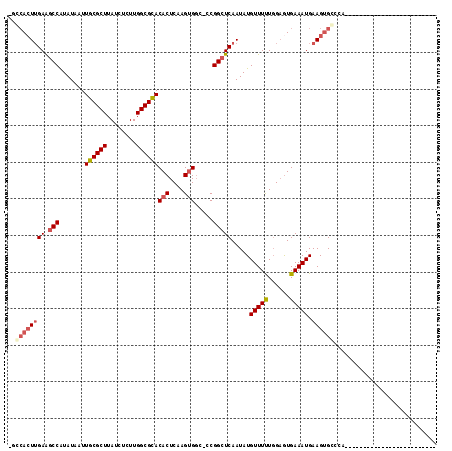
>2L_DroMel_CAF1 16003805 91 - 22407834 AGGGAGUGGAAGCCAUAUAAUUGCGCUUAUCUCUUGGCGCACACUCAAGUGGC-CCGGCUCAAUAUGUUUUUGGAGUGAAAUGAAGUGCCCA------------------------- .(((((...((((((......)).))))..)))))((.((((..(((..(.((-((((..(.....)...)))).)).)..))).)))))).------------------------- ( -23.80) >DroVir_CAF1 165263 110 - 1 --CCACUUGAAUCCAUAUAAUUGCGCUUAUCUCUUGGCGCACACUCAAGUGGCAGCGGCUCAAUAUGUUUCUGGUAUGAAAUGAAGUGGCCAAAAUGCUGCUGCUGUGUCCU----- --.(((((((...........((((((........))))))...)))))))((((((((.......(((((......))))).....(((......))))))))))).....----- ( -32.84) >DroGri_CAF1 159446 96 - 1 --CCACUUGAGGCCAUAUAAUUGCGCUUAUCUCUUGGCGCACACUCAAGUGCCAGCGGCUCAAUAUGUUUCUGGAGUGAAAUGAAGGGG-------------------GCCGCUGCU --.((((((((..........((((((........))))))..)))))))).(((((((((..(.((((((......)))))).)..))-------------------))))))).. ( -39.30) >DroWil_CAF1 153759 104 - 1 GUACACUUGAAGCCAUAUAAUUGCGCUUAUCUCUUGGCGUACACUCAAGUGGCCCCGGCUCAAUAUGUUUUUCGAGUGAAAUGAAGUGCCCAGAAG--UGGG------GCCA----- ...(((((((((.(((((...((((((........))))))(((....)))((....))...)))))..))))))))).......(.((((.....--..))------))).----- ( -30.50) >DroAna_CAF1 114555 91 - 1 CGGCACUUGAAGCCGUAUAAUUGCGCUUAUCUCUUGGCGCACACUCAAGUGGG-CCGGCUCAAUAUGUUUUUGGAGUGAAAUGAAGUGCCCA------------------------- .(((((((((.((((......((((((........))))))(((....)))..-.)))))).....(((((......))))).)))))))..------------------------- ( -30.20) >DroPer_CAF1 227115 92 - 1 UGCCACUUGAAGCCAUAUAAUUGCGCUUAUCUCUUGGCGCACACUCAAGAGGCCCCGGCUCAAUAUGUUUUUGGAGUGAAAUGAAGUGCCCA------------------------- ...(((((.(...........((((((........))))))(((((....(((....)))(((.......))))))))...).)))))....------------------------- ( -23.00) >consensus _GCCACUUGAAGCCAUAUAAUUGCGCUUAUCUCUUGGCGCACACUCAAGUGGC_CCGGCUCAAUAUGUUUUUGGAGUGAAAUGAAGUGCCCA_________________________ ..((((((((.(((.......((((((........))))))(((....))).....))))).....(((((......))))).))))))............................ (-18.72 = -19.47 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:12 2006