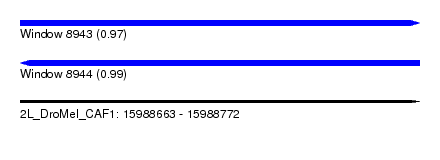
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,988,663 – 15,988,772 |
| Length | 109 |
| Max. P | 0.989881 |

| Location | 15,988,663 – 15,988,772 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
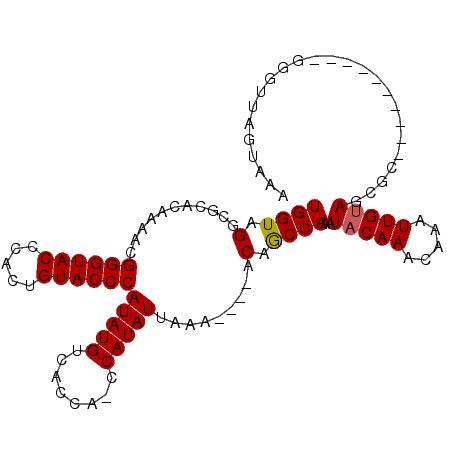
| Reading direction | forward |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -20.40 |
| Energy contribution | -19.73 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15988663 109 + 22407834 UUUACGAACACAUGCAACAGAGUGCUUCAAUGUGUUUGUAUUUGGUUGUGUUUUUUUAUAUGGUUGCUGACAUAUGGGUACAGUAUGUACCCGAUAUGGGCGCCUACCA ..((((((((((((((......)))....)))))))))))..((((.((((......))))(((.(((..(((((((((((.....)))))).))))))))))).)))) ( -34.70) >DroSec_CAF1 164360 95 + 1 UUUACUAACCC----------GCGCUACAAUUUGUUUGUAUUUGGCUGU----UUUAAUAUGGCUGGUGACAUAUGGGUACAGUGGGUACCCGUUUUGUGCGCCUACCA ...........----------((((.((((......(((..(..(((((----......)))))..)..))).((((((((.....)))))))).))))))))...... ( -27.90) >DroSim_CAF1 168043 91 + 1 UUUACUAACCC----------GCGCUACAAUUUGUUUGUAUUUGGCUGU----UUUAAUAUG----GUGACAUAUGGGUACAGUGGGUACCCGUUGUGUGCGCCUACCA ...........----------((((((((((..................----....(((((----....)))))((((((.....)))))))))))).))))...... ( -27.20) >consensus UUUACUAACCC__________GCGCUACAAUUUGUUUGUAUUUGGCUGU____UUUAAUAUGG_UGGUGACAUAUGGGUACAGUGGGUACCCGUUAUGUGCGCCUACCA .....................((((.(((............................(((((........)))))((((((.....))))))....)))))))...... (-20.40 = -19.73 + -0.66)

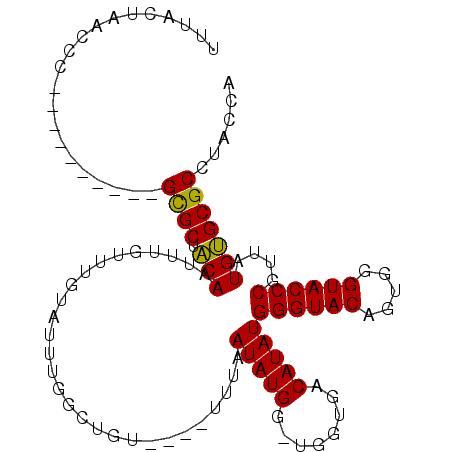
| Location | 15,988,663 – 15,988,772 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -15.55 |
| Energy contribution | -15.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15988663 109 - 22407834 UGGUAGGCGCCCAUAUCGGGUACAUACUGUACCCAUAUGUCAGCAACCAUAUAAAAAAACACAACCAAAUACAAACACAUUGAAGCACUCUGUUGCAUGUGUUCGUAAA ((((..((...(((((.((((((.....)))))))))))...)).))))....................(((.(((((((...(((.....)))..))))))).))).. ( -30.50) >DroSec_CAF1 164360 95 - 1 UGGUAGGCGCACAAAACGGGUACCCACUGUACCCAUAUGUCACCAGCCAUAUUAAA----ACAGCCAAAUACAAACAAAUUGUAGCGC----------GGGUUAGUAAA ((((.((.(.(((....((((((.....))))))...)))).)).))))((((((.----.(.((....(((((.....)))))..))----------)..)))))).. ( -25.40) >DroSim_CAF1 168043 91 - 1 UGGUAGGCGCACACAACGGGUACCCACUGUACCCAUAUGUCAC----CAUAUUAAA----ACAGCCAAAUACAAACAAAUUGUAGCGC----------GGGUUAGUAAA ((((.((((....)...((((((.....))))))....)))))----))((((((.----.(.((....(((((.....)))))..))----------)..)))))).. ( -21.90) >consensus UGGUAGGCGCACAAAACGGGUACCCACUGUACCCAUAUGUCACCA_CCAUAUUAAA____ACAGCCAAAUACAAACAAAUUGUAGCGC__________GGGUUAGUAAA ((((.(...........((((((.....))))))(((((........))))).........).))))..(((((.....)))))......................... (-15.55 = -15.67 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:08 2006