
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,964,956 – 15,965,072 |
| Length | 116 |
| Max. P | 0.846176 |

| Location | 15,964,956 – 15,965,072 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.90 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -25.62 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
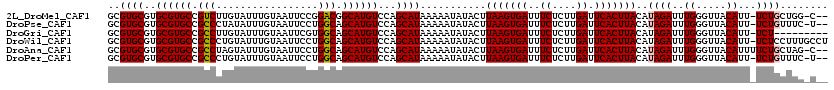
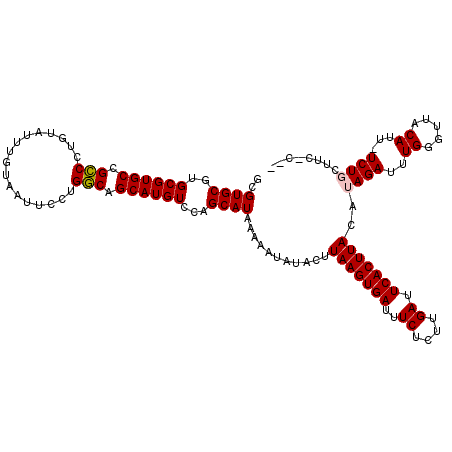
>2L_DroMel_CAF1 15964956 116 + 22407834 GCGUGCGUGCGUGCCGUCUUGUAUUUGUAAUUCCGGACGGCAUGUCCAGCAUAAAAAUAUACUUAAGUGAUUUCUCUUGAUUCACUUACAUAGAUUUGGGUUACAUU-UCUGCUGG-C-- ........(((((((((((.(...........).)))))))))))(((((.............(((((((..((....)).)))))))...(((..((.....))..-))))))))-.-- ( -35.80) >DroPse_CAF1 138883 116 + 1 GCGUGCGUGCGUGCCGCCCUAUAUUUGUAAUUCCUGGCAGCAUGUCCAGCAUAAAAAUAUACUUAAGUGAUUUCUCUUGAUUCACUUACAUAGAUUUGGGUUACAUU-UCUGUUUC-U-- ..((((..((((((.(((.................))).))))))...))))...........(((((((..((....)).))))))).(((((..((.....))..-)))))...-.-- ( -28.73) >DroGri_CAF1 115510 110 + 1 GCGUGCGUGCGUGCCGCCUUGUAUUUGUAAUUCGUGGCAGCAUGUCCAGCAUAAAAAUAUACUUAAGUGAUUUCUCUUGAUUCACUUACAUAGAUUUGGGUUACAUU-UCU--------- ((..((((((.((((((....((....))....))))))))))))...)).............(((((((..((....)).)))))))...(((..((.....))..-)))--------- ( -29.20) >DroWil_CAF1 101256 119 + 1 GCGUGCGUGCGUGCCGCCCUGUAUUUGUAAUUCCUGGCAGCAUGUCCAGCAUAAAAAUAUACUUAAGUGAUUUCUCUUGAUUCACUUACAUAGAUUUGGGUUACAUU-UCUCCUUUGCCU ((((((..((((((.(((.................))).))))))...))))...........(((((((..((....)).)))))))...(((..((.....))..-))).....)).. ( -27.13) >DroAna_CAF1 76023 117 + 1 GCGUGCGUGCGUGCCGCCUAGUAUUUGUAAUUCCUGGCAGCAUGUCCAGCAUAAAAAUAUACUUAAGUGAUUUCUCUUGAUUCACUUACAUAGAUUUGGGUUACAUUUUCUGCUAG-C-- (((.(((....))))))((((((..(((((((((((((.....).)))).....((((.((..(((((((..((....)).)))))))..)).)))))))))))).....))))))-.-- ( -29.00) >DroPer_CAF1 166385 116 + 1 GCGUGCGUGCGUGCCGCCCUGUAUUUGUAAUUCCUGGCAGCAUGUCCAGCAUAAAAAUAUACUUAAGUGAUUUCUCUUGAUUCACUUACAUAGAUUUGGGUUACAUU-UCUGUUUC-U-- ..((((..((((((.(((.................))).))))))...))))...........(((((((..((....)).))))))).(((((..((.....))..-)))))...-.-- ( -28.73) >consensus GCGUGCGUGCGUGCCGCCCUGUAUUUGUAAUUCCUGGCAGCAUGUCCAGCAUAAAAAUAUACUUAAGUGAUUUCUCUUGAUUCACUUACAUAGAUUUGGGUUACAUU_UCUGCUUC_C__ ..((((..((((((.(((.................))).))))))...))))...........(((((((..((....)).)))))))..((((..((.....))...))))........ (-25.62 = -25.68 + 0.06)
| Location | 15,964,956 – 15,965,072 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.90 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -23.67 |
| Energy contribution | -23.12 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
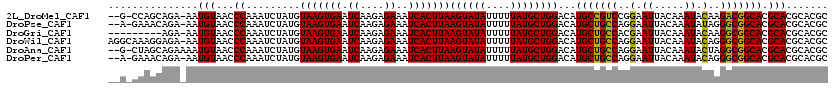
>2L_DroMel_CAF1 15964956 116 - 22407834 --G-CCAGCAGA-AAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCCGUCCGGAAUUACAAAUACAAGACGGCACGCACGCACGC --.-((((((((-((((((.............(((((((.((....))..)))))))..))))))))..))))))...(((((((..(.((.....)).)..)))))))........... ( -29.26) >DroPse_CAF1 138883 116 - 1 --A-GAAACAGA-AAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCAGGAAUUACAAAUAUAGGGCGGCACGCACGCACGC --.-...(((..-..)))...((.........(((((((.((....))..)))))))((((((....))))))))...(((((((.................)))))))........... ( -24.33) >DroGri_CAF1 115510 110 - 1 ---------AGA-AAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCACGAAUUACAAAUACAAGGCGGCACGCACGCACGC ---------(((-((((((.............(((((((.((....))..)))))))..))))))))).(((.(..(.(((((((.................))))))).)..))))... ( -23.19) >DroWil_CAF1 101256 119 - 1 AGGCAAAGGAGA-AAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCAGGAAUUACAAAUACAGGGCGGCACGCACGCACGC .((((....(((-((((((.............(((((((.((....))..)))))))..))))))))).))))(..(.(((((((..(.((.....)).)..))))))).)..)...... ( -25.76) >DroAna_CAF1 76023 117 - 1 --G-CUAGCAGAAAAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCAGGAAUUACAAAUACUAGGCGGCACGCACGCACGC --.-(((((((((((((((.............(((((((.((....))..)))))))..))))))))).))))))...(((((((((............)).)))))))........... ( -29.96) >DroPer_CAF1 166385 116 - 1 --A-GAAACAGA-AAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCAGGAAUUACAAAUACAGGGCGGCACGCACGCACGC --.-...(((..-..)))...((.........(((((((.((....))..)))))))((((((....))))))))...(((((((..(.((.....)).)..)))))))........... ( -25.40) >consensus __G_CAAGCAGA_AAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCAGGAAUUACAAAUACAAGGCGGCACGCACGCACGC ...............(((...((.........(((((((.((....))..)))))))((((((....))))))))...(((((((..(.((.....)).)..))))))).)))....... (-23.67 = -23.12 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:57 2006