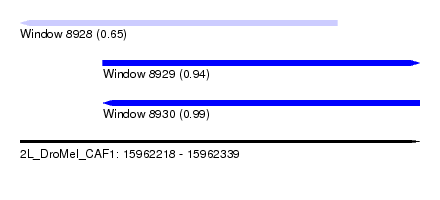
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,962,218 – 15,962,339 |
| Length | 121 |
| Max. P | 0.993026 |

| Location | 15,962,218 – 15,962,314 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 89.51 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -24.41 |
| Energy contribution | -24.97 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15962218 96 - 22407834 GCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCCGAACUCCAUUCCGAGAUCGGAAUAGCCCAGGUCUUCGUUGU ..(((((((.......)))..........(((.((((....))))..)))...((((.(((......))).))))....))))............. ( -29.20) >DroSec_CAF1 137788 96 - 1 GCGGGAGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCCGAACUCCAUUUCGAGAUCGGAAUAGCCCAGGUCUUCGUCGU .((((.(((.......)))..........(((.((((....))))..)))..))))..........(((((((((......)).)))).))).... ( -26.00) >DroSim_CAF1 139935 96 - 1 GCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCCGAACUCCAUUUCGAGAUCGGAAUAGCCCAGGUCUUCGUCGU ..(((((((.......)))..........(((.((((....))))..)))...((((.(((......))).))))....))))............. ( -29.20) >DroEre_CAF1 124877 96 - 1 GCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCCGAACUCCAUUUCGAGAUCGGAAUAGCCCAGGUCGUCGUCAU ..(((((((.......)))..........(((.((((....))))..)))...((((.(((......))).))))....))))............. ( -29.20) >DroYak_CAF1 119913 95 - 1 GCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACC-CGAACUCCAUUUCGAGAUCGGAAUAGCCCAGGUCGUCGUCAU ..(((((((.......)))..........(((.((((....))))..)))..(-(((.(((......))).))))....))))............. ( -29.20) >DroAna_CAF1 73623 82 - 1 ACGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCUAAACUCCAUUUGGUGCUCAGGUUGGCC-------------- ..(((((((.......))).........((((.((((....))))((((((........)))))))))))))).((....))-------------- ( -24.70) >consensus GCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCCGAACUCCAUUUCGAGAUCGGAAUAGCCCAGGUCUUCGUCGU ..(((((((.......)))..........(((.((((....))))..)))...((((.(((......))).))))....))))............. (-24.41 = -24.97 + 0.56)



| Location | 15,962,243 – 15,962,339 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.92 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -16.21 |
| Energy contribution | -16.13 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15962243 96 + 22407834 U--------CUCG-GAAUGGAGUUCGGGGUCCAUUGCGGAUGUCCGCGUGGCAAUUUUAAUGCAAUUUUUAUGCGCCCGCUU-UCGGCUUCCGAUUUUU-UAUUUAA-------- .--------.(((-((((.(((..(((((.((((.((((....))))))))).........(((.......))).))))..)-)).).)))))).....-.......-------- ( -31.20) >DroVir_CAF1 95256 85 + 1 C--------CUCGAAAAUGG-------AGUCCAUUGCGGAUAUCCGCACGGCUAUUUUAAUGCAAUUUUUAUGCGCUCGUU--UUGGCUUCCUAUUUUCU-----UA-------- .--------...((((((((-------((.(((..((((....))))(((((.........(((.......))))).))).--.))).)))).)))))).-----..-------- ( -22.10) >DroPse_CAF1 136042 104 + 1 UGGCACCCACUCU-CUAUGGGGCUCAGGGUCCAUUGCGGAUGUCCGCGUGGCAAUUUUAAUGCAAUUUUUAUGCGCCCGUUU-UUGGCUUCCGAUUUUU-UAUUUCA-------- .(((.((((....-...)))))))..((((((((.((((....))))))))..........(((.......)))))))....-((((...)))).....-.......-------- ( -26.70) >DroGri_CAF1 111777 86 + 1 ------------GGAAAUGG-------AGUCCAUUGCGGAUAUCCGCACAGCAUUUUUAAUGCAAUUUUUAUGCGCUCGUU--UUGGCUUCCUAUUUUCGUAUUUUA-------- ------------((((((((-------((.(((.(((((....)))))..((((.....))))..................--.))).)))).))))))........-------- ( -23.10) >DroAna_CAF1 73634 105 + 1 G--------CACC-AAAUGGAGUUUAGGGUCCAUUGCGGAUGUCCGCGUGGCAAUUUUAAUGCAAUUUUUAUGCGCCCGUUUUUUGGCUUCCGAUCUUU-UAUUUCGUUCUCUCA .--------....-.((((((((..((((((....((((....))))..((..........(((.......)))(((........)))..)))))))).-.))))))))...... ( -25.00) >DroPer_CAF1 163546 104 + 1 UGGCACCCACUCU-CUAUGGGGCUCAGGGUCCAUUGCGGAUGUCCGCGUGGCAAUUUUAAUGCAAUUUUUAUGCGCCCGUUU-UUGGCUUCCGAUUUUU-UAUUUCA-------- .(((.((((....-...)))))))..((((((((.((((....))))))))..........(((.......)))))))....-((((...)))).....-.......-------- ( -26.70) >consensus U________CUCG_AAAUGG_G_UCAGGGUCCAUUGCGGAUGUCCGCGUGGCAAUUUUAAUGCAAUUUUUAUGCGCCCGUUU_UUGGCUUCCGAUUUUU_UAUUUCA________ ..................(((.....((((.((((((((....)))))..(((.......))).......))).))))(((....))).)))....................... (-16.21 = -16.13 + -0.08)


| Location | 15,962,243 – 15,962,339 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.92 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -15.94 |
| Energy contribution | -16.13 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15962243 96 - 22407834 --------UUAAAUA-AAAAAUCGGAAGCCGA-AAGCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCCGAACUCCAUUC-CGAG--------A --------.......-.....((((((...((-...((((.(((.......)))..........(((.((((....))))..)))..))))...))..)))-))).--------. ( -27.90) >DroVir_CAF1 95256 85 - 1 --------UA-----AGAAAAUAGGAAGCCAA--AACGAGCGCAUAAAAAUUGCAUUAAAAUAGCCGUGCGGAUAUCCGCAAUGGACU-------CCAUUUUCGAG--------G --------..-----.((((((.(((..(((.--.(((.(((((.......)))((....)).)))))((((....))))..)))..)-------))))))))...--------. ( -24.50) >DroPse_CAF1 136042 104 - 1 --------UGAAAUA-AAAAAUCGGAAGCCAA-AAACGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCUGAGCCCCAUAG-AGAGUGGGUGCCA --------.......-.....((((...(((.-.....((((((.......))).........)))..((((....))))..)))...))))(((((((..-...))))).)).. ( -27.80) >DroGri_CAF1 111777 86 - 1 --------UAAAAUACGAAAAUAGGAAGCCAA--AACGAGCGCAUAAAAAUUGCAUUAAAAAUGCUGUGCGGAUAUCCGCAAUGGACU-------CCAUUUCC------------ --------...............(((..(((.--.......(((.......))).............(((((....))))).)))..)-------))......------------ ( -19.30) >DroAna_CAF1 73634 105 - 1 UGAGAGAACGAAAUA-AAAGAUCGGAAGCCAAAAAACGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCUAAACUCCAUUU-GGUG--------C ........(((....-.....)))...(((........)))(((.......))).........((((.((((....))))((((((........)))))))-))).--------. ( -23.40) >DroPer_CAF1 163546 104 - 1 --------UGAAAUA-AAAAAUCGGAAGCCAA-AAACGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCUGAGCCCCAUAG-AGAGUGGGUGCCA --------.......-.....((((...(((.-.....((((((.......))).........)))..((((....))))..)))...))))(((((((..-...))))).)).. ( -27.80) >consensus ________UGAAAUA_AAAAAUCGGAAGCCAA_AAACGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCUGA_C_CCAUUU_CGAG________A .......................((..(((........)))(((.......)))..........))..((((....)))).((((..........))))................ (-15.94 = -16.13 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:54 2006