
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,937,622 – 15,937,764 |
| Length | 142 |
| Max. P | 0.865132 |

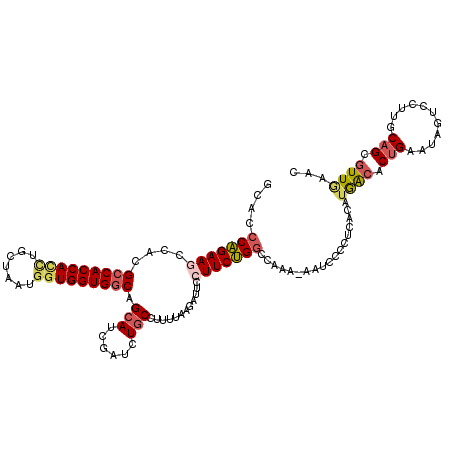
| Location | 15,937,622 – 15,937,741 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -25.49 |
| Energy contribution | -26.13 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
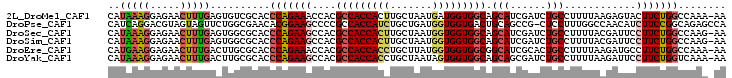
>2L_DroMel_CAF1 15937622 119 - 22407834 GCACCCAGAAACCACGCCACCACUUGCUAAUGAUGGUGGCAGCAUCGAUCUGCCUUUUAAGAGUACUUCUGGCCAAA-AAUCCCCUCACAUGACACUGAAUAGUCCUUGCAGCGUUGAAC (((.((((((...((((((((((........).))))))).(((......))).........))..)))))).....-.............(((........)))..))).......... ( -26.00) >DroPse_CAF1 107046 119 - 1 GAACACGGAAGCCCCGCCACCAUCUGCUGAUGGUGGUGACUGCAGCCG-CUCCUUUGGCCAACAUCUUCCGGCAGAGCCAUCCCCGGACAAAGCGCUGAAUGGUCCGAGCCGCCUUGAAC ......(((((...((((((((((....))))))))))......((((-......))))......)))))(((.(..((((...(((........))).))))..)..)))......... ( -40.70) >DroSec_CAF1 113203 119 - 1 GCACCCAGAAGCCACGCCACCACUUGCUAAUGGUGGUGGCAGCAUCGAUCUGCCUUUUACGAUUCCUUCUGGCCAAG-AAUCCCCUCACUUGACACUGAAUAGUCCUUGCAGCGUUAAAC ....(((((((....(((((((((.......))))))))).(((......)))............))))))).....-...........(((((.(((...........))).))))).. ( -33.80) >DroSim_CAF1 116586 119 - 1 GCACCCAGAAGCCACGCCACCACUUGCUAAUGGUGGUGGCAGCAUCGAUCUGCCUUUUACGAUUCCUUCUGGCCAAG-AAUCCCCUCACAUGACUCUGAAUAGUCCUUGCAGCGUUGAAC (((.(((((((....(((((((((.......))))))))).(((......)))............))))))).....-.............((((......))))..))).......... ( -33.60) >DroEre_CAF1 99538 119 - 1 GCACCCAGAAACCACGCCACCACCUGCUUAUGGUGGUGGCGGCAUCGCACUGCCUUUUAAGAUGCCUUCUGGCCAAA-AAUCCCCUCACAUAACGCUGAACGGUCCUUGCAGCGUUGAAG ....((((((.....(((((((((.......)))))))))((((((..............)))))))))))).....-............((((((((...........))))))))... ( -45.84) >DroYak_CAF1 95550 119 - 1 GCACCCAGAAGCCACGCCACCACCUGCUAAUAGUGGUGGCAGCAGCGAUCUGCCUUUUAAGAUUCCUUCUGGUCAAA-AACCCCCUCACAUGACACUGAACGGUCCUUGCAGCGUUGAAC ((..(((((((....((((((((.........)))))))).((((....))))............))))))).(((.-.(((...(((........)))..)))..)))..))....... ( -33.90) >consensus GCACCCAGAAGCCACGCCACCACCUGCUAAUGGUGGUGGCAGCAUCGAUCUGCCUUUUAAGAUUCCUUCUGGCCAAA_AAUCCCCUCACAUGACACUGAAUAGUCCUUGCAGCGUUGAAC ....(((((((....(((((((((.......))))))))).(((......)))............)))))))..................((((.(((...........))).))))... (-25.49 = -26.13 + 0.64)


| Location | 15,937,662 – 15,937,764 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -27.02 |
| Energy contribution | -26.83 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.723055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15937662 102 + 22407834 UU-UUUGGCCAGAAGUACUCUUAAAAGGCAGAUCGAUGCUGCCACCAUCAUUAGCAAGUGGUGGCGUGGUUUCUGGGUGCGACACUCAAAGUUCUCCUUUAUG .(-(((((((((((.(((........((((......))))((((((((.........))))))))))).).)))))(((...)))))))))............ ( -27.70) >DroPse_CAF1 107086 102 + 1 UGGCUCUGCCGGAAGAUGUUGGCCAAAGGAG-CGGCUGCAGUCACCACCAUCAGCAGAUGGUGGCGGGGCUUCCGUGUUCGCCAGAACUACUACGUCCUGAUG (((((..(((....)..)).))))).(((((-(((..((..((.((((((((....)))))))).)).))..))))((((....)))).......)))).... ( -39.70) >DroSec_CAF1 113243 102 + 1 UU-CUUGGCCAGAAGGAAUCGUAAAAGGCAGAUCGAUGCUGCCACCACCAUUAGCAAGUGGUGGCGUGGCUUCUGGGUGCGCCACUCAAAGUUCUCCUUUAUG ..-........((((((.........(((((.......))))).((((((((....)))))))).(((((..........))))).........))))))... ( -32.30) >DroSim_CAF1 116626 102 + 1 UU-CUUGGCCAGAAGGAAUCGUAAAAGGCAGAUCGAUGCUGCCACCACCAUUAGCAAGUGGUGGCGUGGCUUCUGGGUGCGCCACUCAAAGUUCUCCUUUAUG ..-........((((((.........(((((.......))))).((((((((....)))))))).(((((..........))))).........))))))... ( -32.30) >DroEre_CAF1 99578 102 + 1 UU-UUUGGCCAGAAGGCAUCUUAAAAGGCAGUGCGAUGCCGCCACCACCAUAAGCAGGUGGUGGCGUGGUUUCUGGGUGCGCAAGUCAAAGUUCUCCUUCAUG .(-((((.(((((((.(((.......((((......))))(((((((((.......)))))))))))).)))))))..((....)))))))............ ( -42.10) >DroYak_CAF1 95590 102 + 1 UU-UUUGACCAGAAGGAAUCUUAAAAGGCAGAUCGCUGCUGCCACCACUAUUAGCAGGUGGUGGCGUGGCUUCUGGGUGCGCAAGUCAAAGUUCUCCUUUAUG .(-((((.(((((((...........(((((....)))))(((((((((.......)))))))))....)))))))..((....)))))))............ ( -37.80) >consensus UU_CUUGGCCAGAAGGAAUCGUAAAAGGCAGAUCGAUGCUGCCACCACCAUUAGCAAGUGGUGGCGUGGCUUCUGGGUGCGCCACUCAAAGUUCUCCUUUAUG ........(((((((...........((((......))))((((((((.........))))))))....)))))))........................... (-27.02 = -26.83 + -0.19)

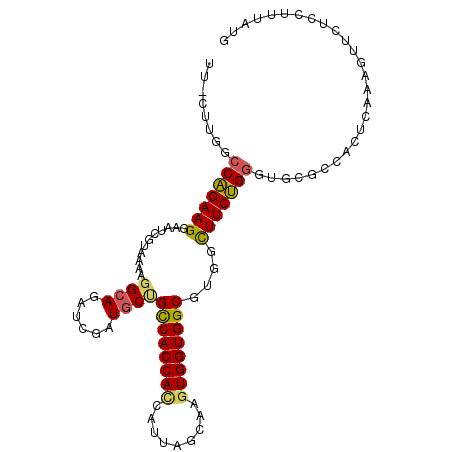

| Location | 15,937,662 – 15,937,764 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -24.61 |
| Energy contribution | -25.20 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15937662 102 - 22407834 CAUAAAGGAGAACUUUGAGUGUCGCACCCAGAAACCACGCCACCACUUGCUAAUGAUGGUGGCAGCAUCGAUCUGCCUUUUAAGAGUACUUCUGGCCAAA-AA ..(((((.....)))))..........((((((...((((((((((........).))))))).(((......))).........))..)))))).....-.. ( -24.20) >DroPse_CAF1 107086 102 - 1 CAUCAGGACGUAGUAGUUCUGGCGAACACGGAAGCCCCGCCACCAUCUGCUGAUGGUGGUGACUGCAGCCG-CUCCUUUGGCCAACAUCUUCCGGCAGAGCCA ....(((((......)))))(((.....((((((...((((((((((....))))))))))......((((-......))))......)))))).....))). ( -41.60) >DroSec_CAF1 113243 102 - 1 CAUAAAGGAGAACUUUGAGUGGCGCACCCAGAAGCCACGCCACCACUUGCUAAUGGUGGUGGCAGCAUCGAUCUGCCUUUUACGAUUCCUUCUGGCCAAG-AA ...((((((((...(((((((((..........)))))(((((((((.......)))))))))....))))))).)))))..........(((.....))-). ( -34.00) >DroSim_CAF1 116626 102 - 1 CAUAAAGGAGAACUUUGAGUGGCGCACCCAGAAGCCACGCCACCACUUGCUAAUGGUGGUGGCAGCAUCGAUCUGCCUUUUACGAUUCCUUCUGGCCAAG-AA ...((((((((...(((((((((..........)))))(((((((((.......)))))))))....))))))).)))))..........(((.....))-). ( -34.00) >DroEre_CAF1 99578 102 - 1 CAUGAAGGAGAACUUUGACUUGCGCACCCAGAAACCACGCCACCACCUGCUUAUGGUGGUGGCGGCAUCGCACUGCCUUUUAAGAUGCCUUCUGGCCAAA-AA ...((((.....))))...........((((((.....(((((((((.......)))))))))((((((..............)))))))))))).....-.. ( -37.94) >DroYak_CAF1 95590 102 - 1 CAUAAAGGAGAACUUUGACUUGCGCACCCAGAAGCCACGCCACCACCUGCUAAUAGUGGUGGCAGCAGCGAUCUGCCUUUUAAGAUUCCUUCUGGUCAAA-AA ..(((((.....)))))..........(((((((....((((((((.........)))))))).((((....))))............))))))).....-.. ( -30.90) >consensus CAUAAAGGAGAACUUUGACUGGCGCACCCAGAAGCCACGCCACCACCUGCUAAUGGUGGUGGCAGCAUCGAUCUGCCUUUUAAGAUUCCUUCUGGCCAAA_AA ..(((((.....)))))..........(((((((....(((((((((.......))))))))).(((......)))............)))))))........ (-24.61 = -25.20 + 0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:50 2006