| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,927,895 – 15,927,993 |
| Length | 98 |
| Max. P | 0.611173 |

| Location | 15,927,895 – 15,927,993 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -17.72 |
| Energy contribution | -18.05 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15927895 98 + 22407834 GGUUAAGUAUGUGGGUUAAUUGCAUAAGUUAAAGCCGAAUGAAGCGGCUAUUUAAAUUAUUGCACACCACGUGUGCCCCUCAGAU--GGGCAAA------AAAAGG .......(((((((......(((((((.(((((((((.......)))))..)))).))).))))..)))))))(((((.......--)))))..------...... ( -26.60) >DroSec_CAF1 103043 97 + 1 GGUUAAGUAUGUGGGUUAAUUGCAUAAGUUAAAGCCGAAUGAAGCGGCUAUUUAAAUUAUUGCAUACCGCGUGUGUCCCUCAGUU--GGGCAAA-------AAAGG .......(((((((......(((((((.(((((((((.......)))))..)))).))).))))..)))))))((.(((......--)))))..-------..... ( -24.80) >DroSim_CAF1 105057 97 + 1 GGUUAAGUAUGUGGGUUAAUUGCAUAAGUUAAAGCCGAAUGAAGCGGCUAUUUAAAUUAUUGCAUACCACGUGUGUCCCUCAGUU--GGGCAAA-------AAAGG .......(((((((......(((((((.(((((((((.......)))))..)))).))).))))..)))))))((.(((......--)))))..-------..... ( -25.20) >DroEre_CAF1 89899 103 + 1 GGUUAAGCAUGUGGGUUAAUUGCAUAGGUUAAAGCCGAGUGAAGCGGCGAUUUAAAUUAUUGCA-ACCACGUGUGUCCCUGAGAU--GGACAAAAGUGCAAAAAAG ......((((((((.....((((((((.((((.((((.......))))...)))).))).))))-)))))))))((((.......--))))............... ( -26.80) >DroYak_CAF1 84505 96 + 1 GGUUAAGCAUGUGGGUUAAUUGCACAAGUUAAAGCCGAAUGAAGCCGCGAUUUAAAUUAUUGCAUACCACGUGUGUCCCUGAGAU--GG--------GCAGAAAAG ......((((((((.((((((.....))))))..............(((((.......)))))...))))))))(.(((......--))--------))....... ( -22.70) >DroAna_CAF1 27401 92 + 1 GG-CAGGCAUAUGGGUUAAUUGCAUAAAUUAAAGCCGCAUGAAGCGGCUAUUUAAAUUAUUUUCCACCACGUGU--CACGGAACUACCGGCAAA-------A---- .(-(.((((..(((.((((((.....))))))((((((.....)))))).................)))..)))--).(((.....)))))...-------.---- ( -28.10) >consensus GGUUAAGCAUGUGGGUUAAUUGCAUAAGUUAAAGCCGAAUGAAGCGGCUAUUUAAAUUAUUGCAUACCACGUGUGUCCCUCAGAU__GGGCAAA_______AAAGG ((....((((((((......(((((((.(((((((((.......))))..))))).))).))))..))))))))...))........................... (-17.72 = -18.05 + 0.33)


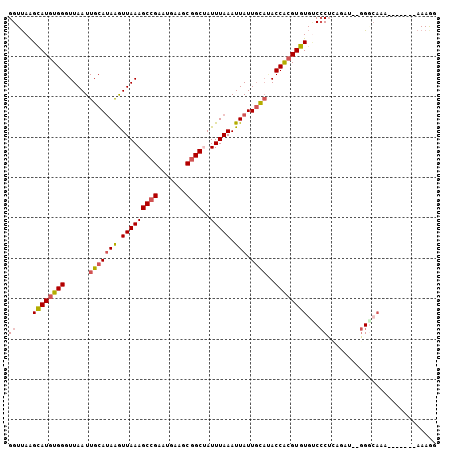
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:45 2006